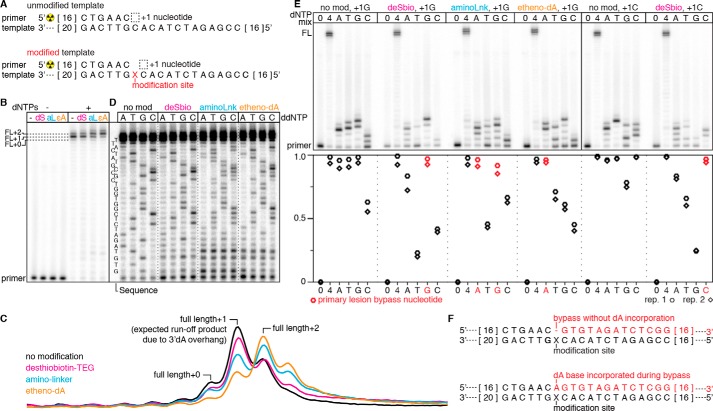
Figure 2.
Characterization of Sulfolobus DNA Pol IV-mediated DNA modification bypass. A, overview of the primer extension reaction on an unmodified and internally modified template. The length of DNA nucleotides that are not shown is indicated by the numbers in brackets at the ends of the sequence. B, characterization of full-length (FL) primer extension products for unmodified (−) and desthiobiotin–TEG (dS)–modified, amino linker (aL)–modified, and etheno-dA (ϵA)–modified templates. The primary runoff product is FL+1 because of untemplated incorporation of a 3′dA overhang; some runoff products contain a second overhang nucleotide (23). The amino linker and etheno-dA modifications increase the occurrence of FL+2, suggesting incorporation of a dNTP opposite the modification site upon lesion bypass. C, raw intensity traces of the full-length products from B. D, ddNTP sequencing ladders for unmodified and desthiobiotin–TEG (deSbio)–modified, amino linker (aminoLnk)–modified, and etheno-dA–modified templates. In agreement with B, bypass of the desthiobiotin–TEG modification yields a sequencing ladder that resembles the unmodified control. In contrast, the amino linker and etheno-dA templates yield a mixed sequencing ladder that suggests incorporation of an additional base during modification bypass. For each modified oligonucleotide, ddNTP truncation products within four to five nucleotides of the modification site were obscured by short products that were not fully extended. E, primer extension reactions for unmodified and modified DNA templates performed in the absence of dNTPs (0), with all dNTPs (4), or with only one dNTP (A, T, G, or C). The unmodified control and desthiobiotin–TEG reactions were performed with the sequences shown in A (+1G) and with a template in which the +1 nucleotide is a C (+1C). Quantification of primer extension is shown in the plot below; red points indicate individual dNTP conditions that enable high-efficiency modification bypass. F, schematic of primer extension products resulting from bypass without dA incorporation (top) and bypass with dA incorporation opposite the modification site (bottom). B–D are representative of two independent replicates. The data in B are controls that were performed alongside the reactions in D; the solid line between these two panels indicates a gel splice site used to facilitate clear visualization of the sequencing ladders in D. In E, n = 2.