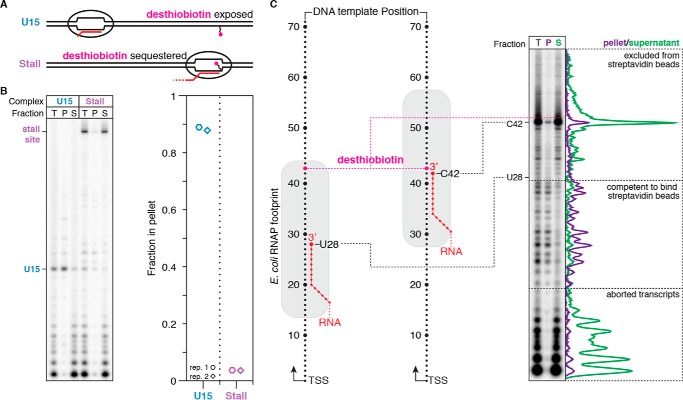
Figure 5.
Characterization of desthiobiotin sequestration by E. coli RNAP. A, overview of the desthiobiotin sequestration experiment in B. If RNAP is positioned at U15, then the desthiobiotin–TEG modification is predicted to be exposed and capable of binding streptavidin-coated magnetic beads. If RNAP is positioned at the stall site, then the desthiobiotin–TEG modification is predicted to be sequestered and incapable of binding streptavidin-coated magnetic beads. B, fractionation of internal desthiobiotin–TEG–modified DNA templates with streptavidin-coated magnetic beads in the presence of the TECs described in A. Total reaction (T), pellet (P), and supernatant (S) fractions are shown. The plot shows the fraction of TECs retained in the bead pellet. C, fractionation of internal desthiobiotin–TEG–modified DNA templates with streptavidin-coated magnetic beads in the presence of intermediate TECs. Intermediate TECs were generated by transcription in the presence of chain-terminating 3′-dNTPs. Comparison of P and S fractions indicates that the E. coli RNAP footprint blocks desthiobiotin from binding streptavidin when the nascent RNA 3′ end is 15 nucleotides or less upstream of the modification site. Background-subtracted intensity traces are shown for the pellet and supernatant fractions alongside the gel. All data are from two independent replicates.