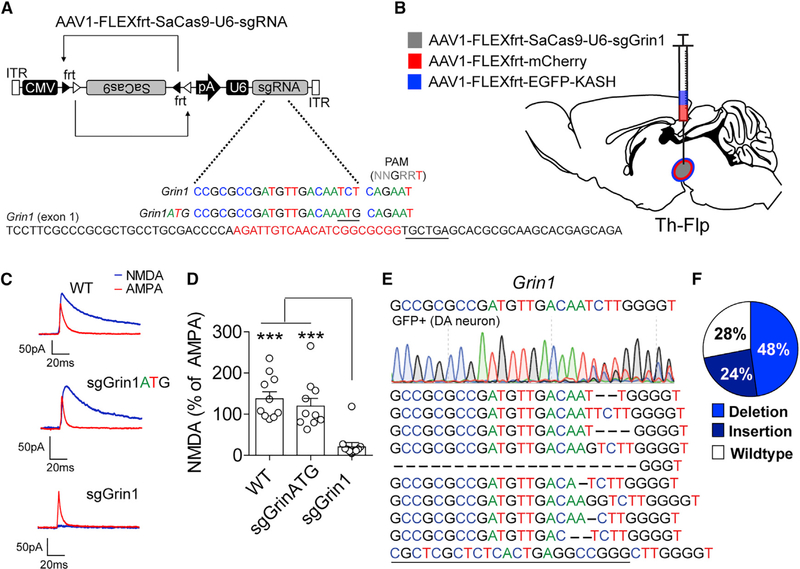
Figure 7. Flp-Dependent CRISPR/SaCas9 Produces Robust Gene Mutagenesis.
(A) Design of AAV1-FLEXfrt-SaCas9-U6-sgGrin1 and control virus AAV1-FLEXfrt-SaCas9-U6-sgGrin1ATG. Exon 1 of Grin1 is denoted with guide sequence highlighted in red; the PAM is underlined.
(B) Illustration of sagittal section with viral injections into the VTA.
(C) Representative traces of NMDA and AMPA currents evoked in dopamine neurons by electrical stimulation in WT mice or mice expressing sgGrin1 or sgGrin1ATG. AMPA current recordings were made in the presence of AP5; NMDA currents were determined by subtracting the AMPA current from the compound current trace recorded in ACSF.
(D) Quantification of the peak NMDA current as a percentage of the peak AMPA current (n = 10 cells/group; one-way ANOVA, F[2, 27] = 15.55, p < 0.0001; Tukey’s multiple-comparisons test, ***p < 0.001).
(E) Sanger sequencing of FACS sorted GFP-positive nuclei from the combined gate of mice co-injected with AAV1-FLEXfrt-SaCas9-U6-sgGrin1 and AAV1-FLEXfrt-EGFP-KASH into the VTA of Th-Flp mice and the most common indels (bottom).
(F) Percentage of targeted deep sequencing reads with wild-type, insertion, deletion, or base changes. Data are presented as mean ± SEM.