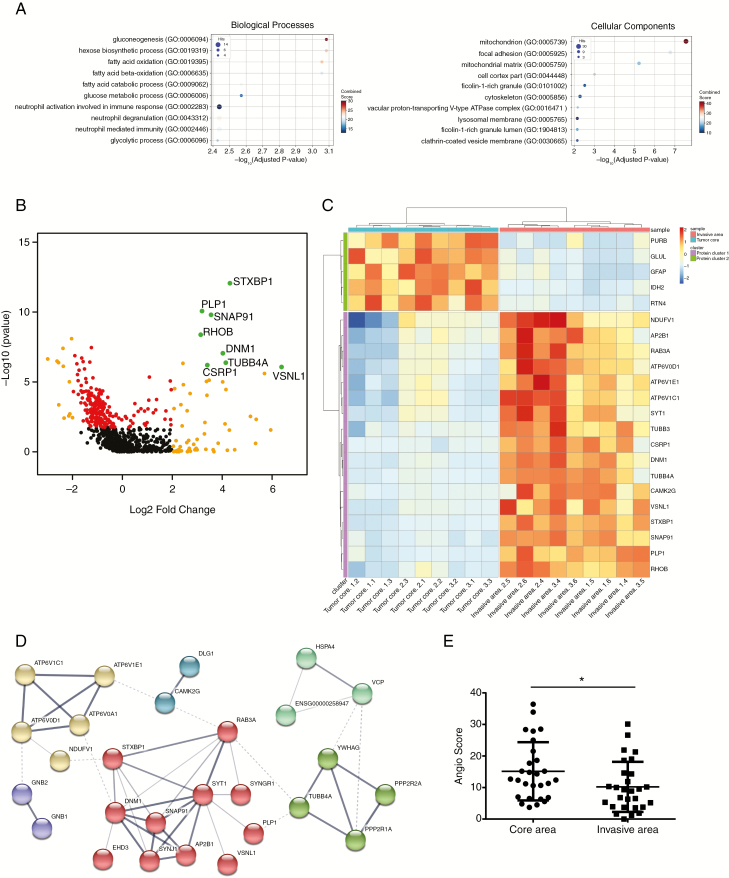
Figure 2.
Protein enrichment of tumor invasive area (A) Gene Set Enrichment Analysis results. Gene Ontology (GO) biological processes (BP) and cellular components (CC) enrichment of the top 152 proteins that are significantly differentially expressed between core and invasive areas (pval adj < 0.01 and |logFC| > 2), and which are represented here for the three tumors. The x-axis represents the negative log10 P-adjusted value. The size of each spot (Ratio) corresponds to the fraction of proteins within our set of proteins that have the corresponding GO function. (B) Graphical representation of quantitative proteomics data for the three P3 tumors. Proteins are ranked in a volcano plot according to P value for technical reproducibility, calculated from a one-tailed paired t-test (−log10 [P value]) (y-axis) and their relative abundance ratio (log ratio) between core and invasive areas (x-axis). The line indicates the P < 0.05 threshold. Off-centered spots are those that vary the most between core and invasive areas. (C) Heatmap of the top 22 of the 152 proteins that are significantly differentially expressed for P3 tumor core and invasive areas (pval adj < 0.01 and |logFC| > 2). (D) Network of human tumor invasive proteins. Each node represents a protein and interactions with medium confidence >0.4 are showed. Proteins are clustered using the Markov Cluster Algorithm and colors represent clusters. Dashed lines represent intercluster edges and width represent edge confidence (medium: >0.4, high: >0.7, and highest: 0.9). Disconnected proteins are removed from the analysis. (E) AngioScore of the top 30 hits from P3 tumor core and invasive areas. Student t-test, * P < .05.