Fig. 1:
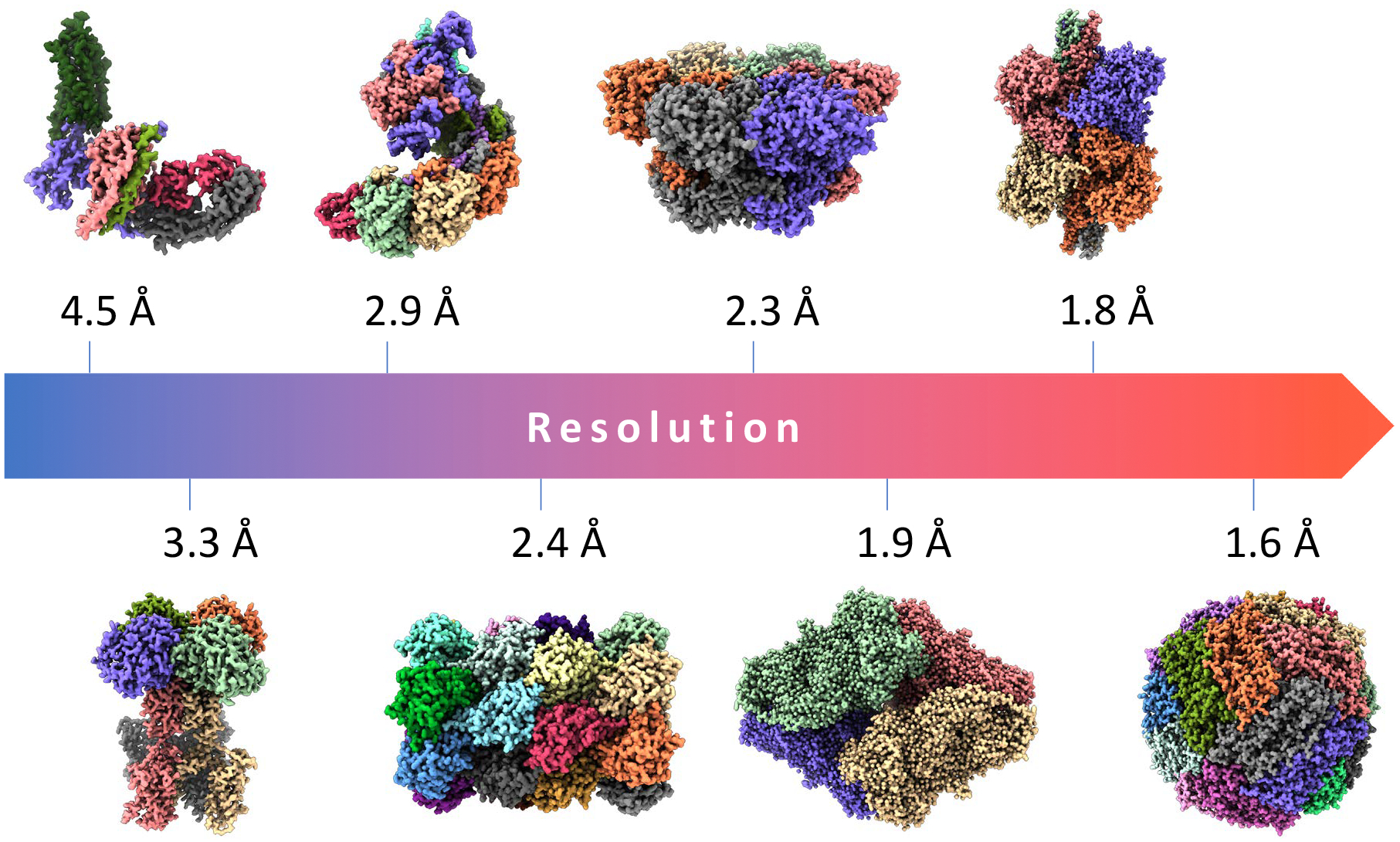
Gallery of important biomedical structures solved by single-particle cryo-EM at increasing resolutions. From left to right (lower to higher resolution): 4.5Å structure of the human rhodopsin receptor bound to an inhibitory G protein [43], a member of the family of G-protein coupled receptors which are the target of ~ 35% of FDA-approved drugs; 3.3Å map of a voltage-activated potassium channel, an integral membrane protein responsible for potassium ion transport [49];2.9Å cryo-EM structure of a CRISPR-Cas Surveillance Complex implicated in gene editing [36];2.4Å map of the T20s proteasome, a complex that degrades unnecessary or damaged proteins by proteolysis (data available from EMPIAR-10025) [1]; 2.3Å structure of human p97 AAA-ATPase, a key mediator of several protein homeostasis processes and a target for cancer [12]; 1.9Å structure of β-galactosidase enzyme in complex with a cell-permeant inhibitor (EMPIAR-10061) [15];1.8Å structure of the conformationally dynamic enzyme glutamate dehydrogenase [50]; and 1.6Å map of human apoferritin, a critical intracellular iron-storage protein (EMPIAR-10200).
