Fig. 4:
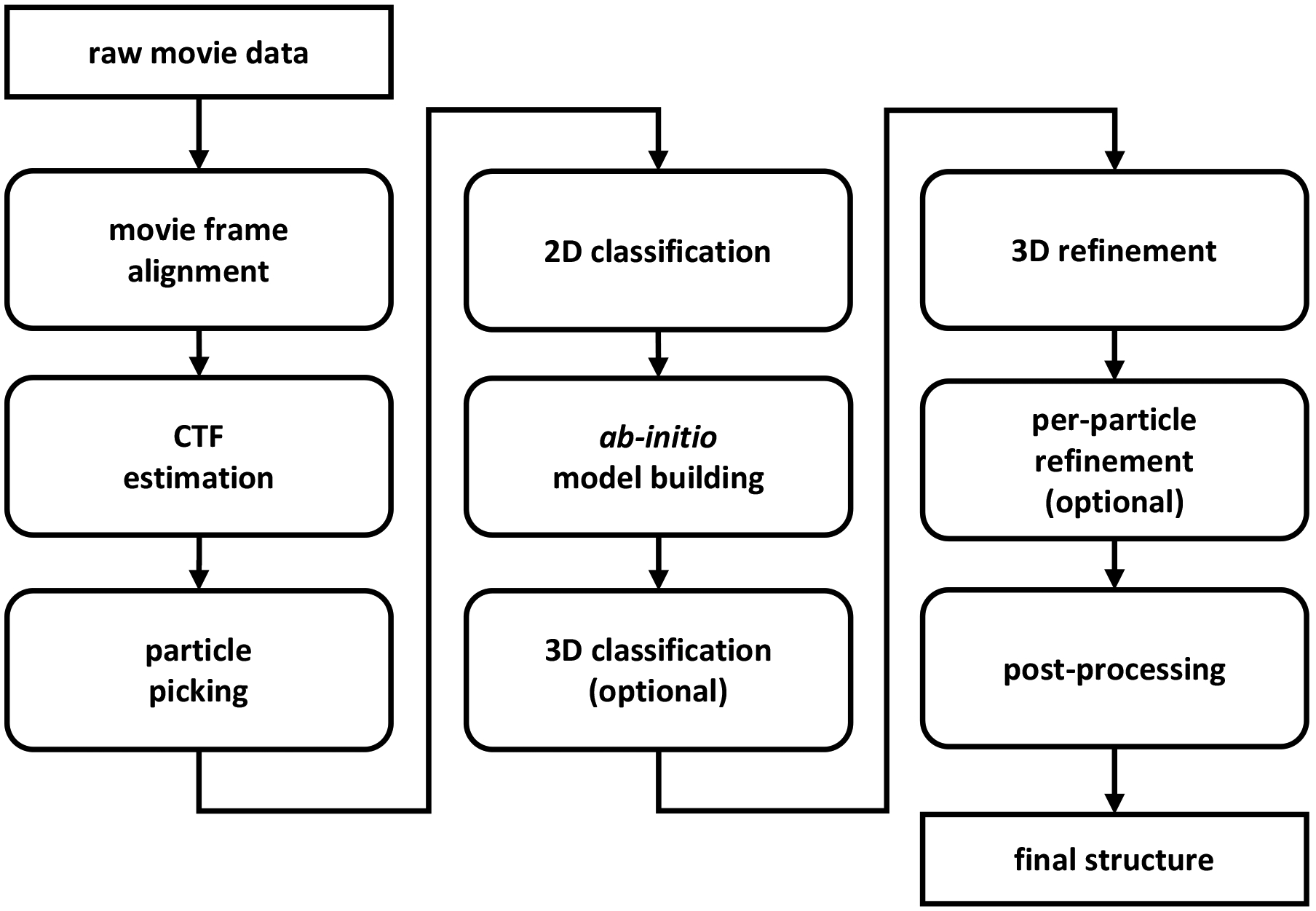
Flowchart diagram showing the computational pipeline required to convert raw movie data into high-resolution structures by single-particle cryo-EM. Raw data is first pre-processed at the movie frame alignment (Section V-A) and CTF estimation (Section V-B) steps followed by particle picking to detect and extract the individual projections from micrographs. Occasionally, particle picking is followed by a particle pruning stage to remove non-informative picked particles (Section V-C). The output of this stage is a set of 2-D images, each (ideally) contains a noisy tomographic projection taken from an unknown viewing direction. Particle images are then classified (Section V-D): the 2-D classes are used to construct ab initio models, as templates for particle picking, to provide a quick assessment of the particles, and for symmetry detection. Then, an initial, ab initio model is built using the 2-D classes or by alternative techniques (Section V-D). If the data contains structural variability or a mix of structures (see Sections II and VII-A), then a 3-D classification step is applied to cluster the projection images into the different structural conformations. Initial models are subjected to 3-D high-resolution refinement (Section IV-A) and an additional per-particle refinement may be applied. Finally, a post-processing stage is employed to facilitate interpretation of structures in terms of atomic models. Different software packages may use slightly different workflows and occasionally some of the steps are applied iteratively. For instance, one can use the 2-D classes to repeat particle picking with more reliable templates. This figure is adapted from [90].
