Figure 1.
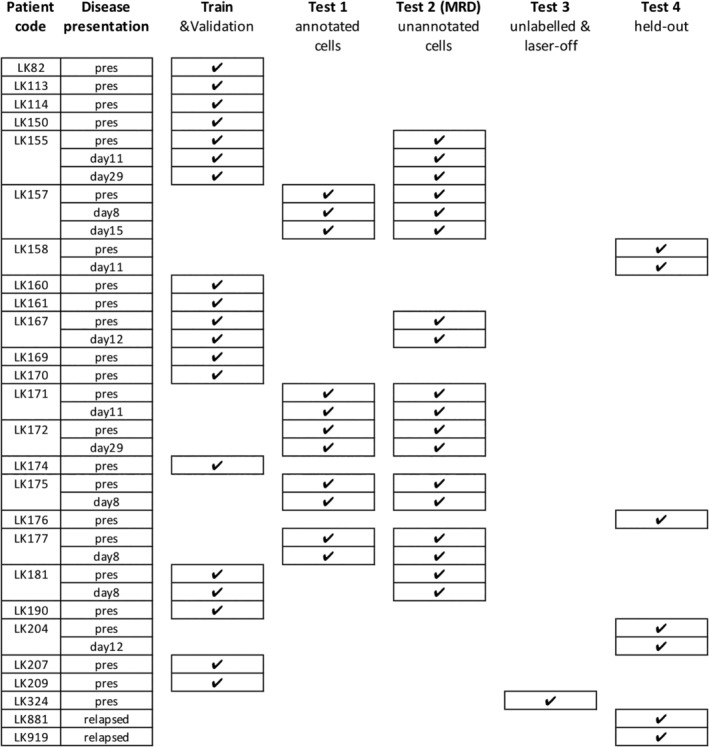
Sample partitioning strategy for training, validation, inference, and avoiding overfitting. Samples were split for training (including validation), testing (Test 1–3) and hold‐out (Test 4). Training/validation set contained pooled data of 19 entries from 15 patients. Samples were collected and measured at the time of presentation (abbreviated as “pres”) and after round(s) of treatments (noted as days after treatment). Test set 1 contained manually gated ground‐truth populations for leukemic blasts, normal lymphocytes and other cell types (Fig. 2A‐C). Test set 2, which contains DAPI‐positive, in‐focus single white blood cells, was designed to validate whether the learned algorithms were able to derive a correct residual disease (MRD) readout, that is, percentage of leukemic cells within the total number of white cells in the bone marrow sample (Fig. 2D). Note: Although some training data and Test set 2 were generated from the same patients, the training sets use a small number of individually annotated healthy/leukemic cells, while Test set 2 presents a large number of unannotated cells. Test set 3 (>200,000 single cells in total) was conducted with stained/unstained samples in a condition with or without laser illumination, confirming that the performance of the trained neural network was not dependent on the presence of bleed‐through fluorescence or lasers (Fig. 2E). Test set 4 was kept held‐out and only unlocked immediately before submission of the manuscript for the final verification of the success of the machine learning models (Fig. 2F).
