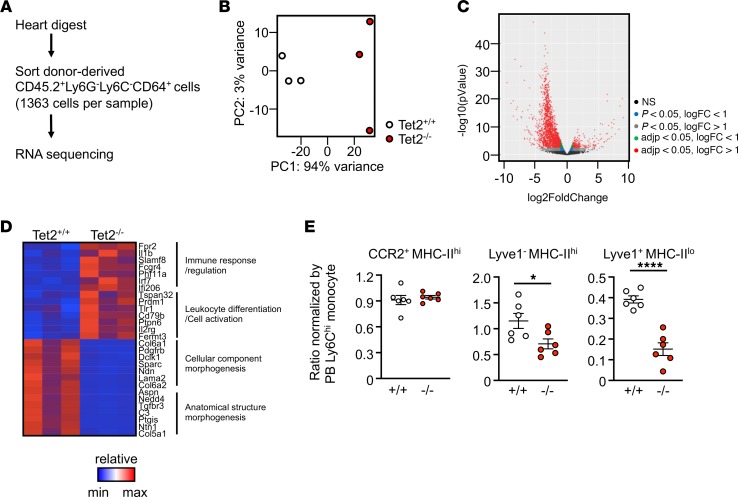
Figure 5. RNA-Seq analysis of Tet2-sufficient and Tet2-deficient donor-derived cardiac macrophages.
(A) Scheme of the study. Donor bone marrow–derived macrophages were defined as the CD45.2+CD64+Ly6G–Ly6C– population. At 8 months after adoptive transfer, 1363 cells were sorted from mice and used for ultralow-input RNA-Seq. (B) Principal component analysis of ultralow-input RNA-Seq data obtained from sorted donor bone marrow–derived cardiac macrophages of Tet2-sufficient and Tet2-deficient mice (n = 3 per genotype). (C) Volcano plots showing the number of genes differentially expressed between cardiac macrophages derived from WT versus Tet2-deficient bone marrow cells. (D) Heatmap of a select group of the most highly differentially expressed genes from the highest ranked annotation categories comparing cardiac macrophages derived from WT versus Tet2-deficient bone marrow cells. (E) Result of donor chimerism of cardiac subpopulations normalized by donor chimerism of peripheral blood Ly6Chi monocytes (n = 6 per genotype). Statistical analysis was performed with 2-tailed unpaired Student’s t test. *P < 0.05, and ****P < 0.0001.