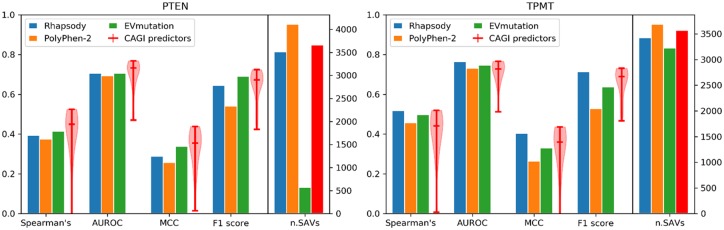
Fig. 7.
Assessment of pathogenicity predictions applied to PTEN and TPMT variants. The level of agreement between computationally predicted pathogenicity scores and experimental abundance scores is shown, based on four metrics (Spearman’s, AUROC, MCC and F1-score). Results are presented for Rhapsody, PolyPhen-2 and EVmutation, represented by bar plots (see also scatter plots in Supplementary Fig. S16), and for CAGI predictors, represented by violin plots showing the distribution, median and range of values. The ‘low-abundance’ and ‘possibly low-abundance’ classes [as reported in (Matreyek et al., 2018)] were considered as ‘deleterious’, and ‘possibly WT-like’ and the ‘WT-like’ classes, as ‘neutral’. The rightmost bars in each plot represent the number of SAVs successfully predicted by each method (in red, the common value for CAGI predictors)