Fig. 6.
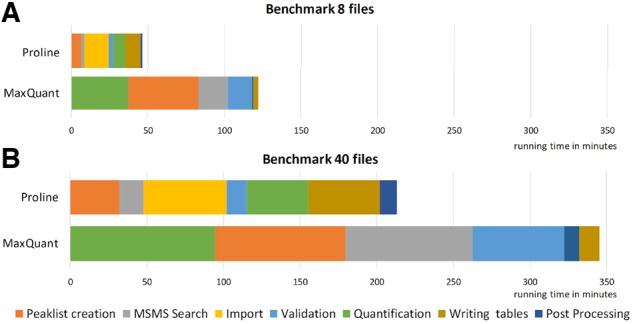
Workflows computation time. Performance of the Mascot–Proline and Andromeda–MaxQuant label-free workflows were compared on two datasets of different sizes. The main steps of the compared workflows are shown in the same color when possible. Time values were taken from the ‘runningTimes.txt’ output file for MaxQuant and from the log files for Proline. (A) Performance observed for a dataset containing eight UPS1-Yeast LC-MS/MS runs: Total processing time was 122 min for MaxQuant and 46 min for Proline (average time per file 15.26 and 5.79 min, respectively). (B) Performance observed for the whole UPS1-Yeast dataset (40 LC-MS/MS runs): Total processing time 346 min for MaxQuant and 214 min for Proline (average time per file: 8.63 and 5.34 min, respectively)
