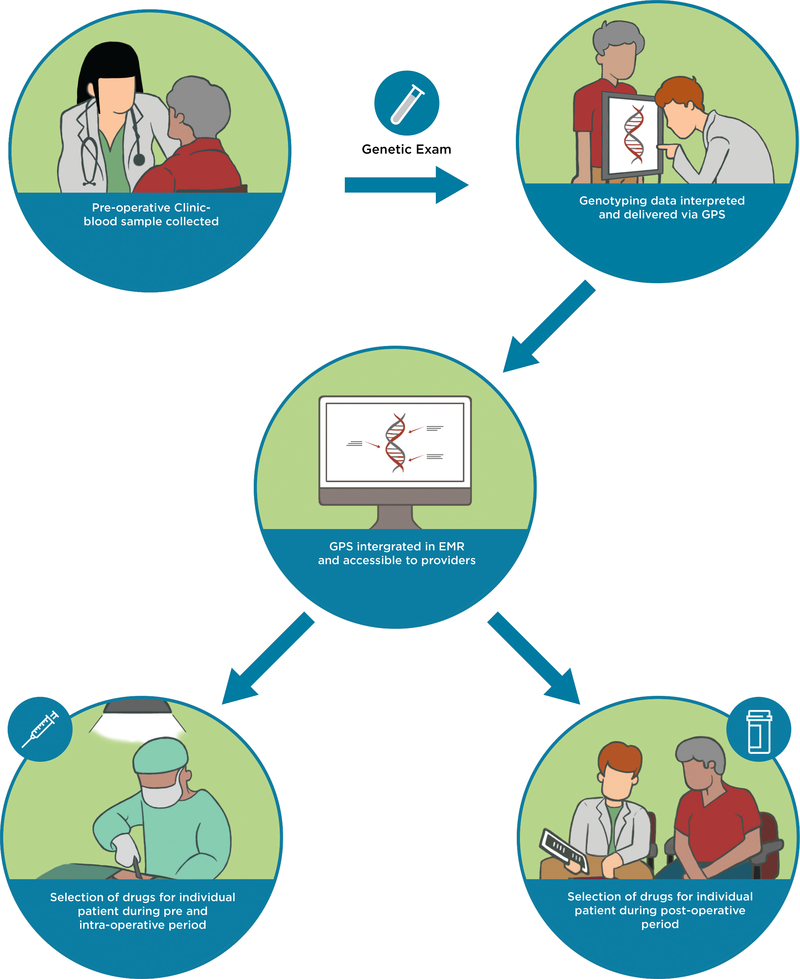
Figure 2.
A model for implementing pharmacogenomics into the current perioperative workflow. Patient exclusion criteria include 1) patients who have undergone, or are being actively considered for, liver or kidney transplantation; 2) patients with known active or prior leukemia; and 3) patients who are unable to understand and give informed consent to participate. During the patients’ pre-operative clinic visit at the APMC, a sample of whole blood will be collected in conjunction with standard-of-care phlebotomy. Genotyping will be carried out in the CLIA-certified and CAP-accredited Clinical Pharmacogenomics Laboratory at The University of Chicago. Raw genotyping data will be interpreted and delivered to providers as clinical decision support summaries through our clinical portal, the GPS, which is available as a link-out within our institutional EMR. Participating providers can use the GPS to guide selection of drugs pre-operatively, intra-operatively, and post-operatively. A clinical research coordinator from the study team will be embedded in the APMC to actively recruit eligible patients. The research coordinator will also be readily available throughout the perioperative period to alert providers about enrolled patients upon admission as well as to help them navigate GPS. This will allow us to integrate ourselves within the perioperative workflow and help us determine key points during the perioperative workflow that need to improve for our implementation program. APMC = Anesthesia Perioperative Medicine Clinic; CLIA = Clinical Laboratory Improvement Amendments; CAP = College of American Pathologists; GPS = Genomic Prescribing System; EMR = electronic medical record.