Abstract
Purpose
Osteosarcoma (OS) is the most common primary bone tumor, with high morbidity in infants and adolescents. Long noncoding RNA LINC00313 has been found to modulate papillary thyroid cancer tumorigenesis and to be dysregulate in lung cancer. However, the role of LINC00313 in OS has not yet been addressed.
Materials and Methods
We evaluated mRNA and protein expression using real-time quantitative PCR and Western blotting. Cell proliferation was evaluated using MTT; apoptosis and autophagy were assessed with flow cytometry, Western blotting, and/or GFP-LC3 assay. Transwell assay was conducted to measure cell migration and invasion. Potential target sites for LINC00313 and miR-342-3p were predicted with starBase v.2.0 and TargetScan Human, and verified using luciferase reporter assay, RNA immunoprecipitation, and RNA pull-down assay. In vivo, xenogeneic tumors were induced with U2OS and MG-63 cells, separately.
Results
LINC00313 was upregulated and miR-342-3p was downregulated in OS tissues and cells. High expression of LINC00313 was associated with shorter overall survival. FOSL2 downregulation and miR-342-3p overexpression suppressed cell proliferation and migratory and invasive abilities while promoting apoptosis and autophagy, all of which were consistent with the effects of LINC00313 knockdown. miR-342-3p, sponged by LINC00313, inversely modulated FOSL2 by targeting MG-63 cells, and FOSL2 expression was positively controlled by LINC00313. LINC00313 knockdown suppressed tumor growth in vivo.
Conclusion
LINC00313 is upregulated in OS, and LINC00313 knockdown plays a vital anti-tumor role in OS cell progression through a miR-342-3p/FOSL2 axis. Our study suggests that LINC00313 may be a novel, promising biomarker for diagnosis and prognosis of OS.
Keywords: LINC00313, miR-342-3p, FOSL2, osteosarcoma (OS)
INTRODUCTION
Osteosarcoma (OS) is a highly malignant, very aggressive bone tumor in children and adolescents.1 It is characterized by early metastasis through blood to other tissues, especially the lungs.2 Several studies have suggested that OS arises from primitive mesenchymal bone-forming cells.3,4,5 Treatment involves standard chemotherapy schemes before and after surgery, and has increased the 5-year survival rate to 60–70%.6 However, the median survival time for these patients is only 23 months, and half of the patients develop drug-resistance and face a high risk of relapse. Current clinical trials of cytotoxicity chemotherapy and targeted agents appear to be able to improve certain OS characteristics, but not overall survival.7 Accordingly, identifying diagnostic and prognostic markers and therapeutic targets for the management of patients with OS is needed.
Increasing evidence indicates that many of the genomic mutations in cancer reside in regions that do not encode proteins and are often transcribed into long non-coding RNAs (lncRNAs).8 LncRNAs are in the range from 200 nt to 100 kb9 and exist both in the cytoplasm to regulate mRNA translation and stability, to act as miRNAs precursors, or to compete with endogenous RNAs (ceRNAs) to modulate miRNAs distributions and in the nucleus to interact with promoters in epigenetic regulation.6 As discussed in a previous review, lncRNAs are frequently expressed in human cancers, promoting tumor development, progression, and metastasis.10 LINC00313 is a novel lncRNA and has been found to be upregulated, to be associated with the poor prognosis of lung cancer (LC), and to modulate papillary thyroid cancer (PTC) tumorigenesis.11,12 With a lack of research on LINC00313 in various tumors, the aim of this study was to investigate the role of LINC00313 in OS.
MicroRNAs (miRNAs), a single-stranded non-coding RNA containing 22–24 nt, regulate the occurrence, development, and prognosis of tumors and cancers. MiR-342-3p has been found to be consistently dysregulated in various diseases, including diabetic kidney disease, colorectal carcinoma, and malignant melanoma.13,14,15 In addition, miR-342-3p has been shown to universally act as a tumor suppressor gene and to be downregulated in cancers. Previous research indicated that miR-342-3p exerts inhibition effects on OS cell proliferation, migration, and invasion.16 Although much is understood about how miR-342-3p regulates tumorigenesis, how this master regulator is itself regulated remains largely unknown.
Fos-like antigen 2 (FOSL2), also named FRA-2, belongs to the activator protein 1 (AP-1) transcription factor family, which includes the various isoforms of Fos and Jun.17 FOSL2 is widespread in human tissues and plays an important role in cancer metastasis.18 Studies have shown that overexpression of FOSL2 is associated with higher migratory and invasive abilities in breast cancer, colon cancer, and hepatocellular carcinoma.18,19,20 Very recent data have indicated that FOSL2 participates in OS cell proliferation, migration, and invasion.21 Moreover, it has been reported that several miRNAs play a regulatory role by directly inhibiting target FOSL2 in cancers.
In this study, we examined differences in the expressions of LINC00313 and miR-342-3p between OS tissues/cells and normal tissues/cells, and explored the biological functions of LINC00313 and miR-342-3p in proliferation, apoptosis, migration, invasion, and autophagy in OS cell lines (U2OS and MG-63). Meanwhile, we investigated the relationship between LINC00313 and its target gene miR-342-3p, and determined that LINC00313 knockdown confers good prognosis and plays a vital anti-tumor role in suppressing cell proliferation, migration, and invasion and in promoting cell apoptosis and autophagy through a miR-342-3p/FOSL2 axis.
MATERIALS AND METHODS
Acquirement of OS tissue
With approval from the Research Ethics Committee of the Sixth Affiliated Hospital of Xinjiang Medical University and with written informed consent from primary OS patients and/or their guardians, a total of 87 tissue samples were obtained from 2013 to 2017, along with 24 adjacent normal tissues collected at the time of operation. Prior to surgery, none of these OS patients received any anti-cancer treatment. The detail clinical features of the 87 OS patients are presented in Table 1. All tissue samples were immediately stored in liquid nitrogen.
Table 1. Correlation between LINC00313 Expression and Clinicopathological Parameters of Osteosarcoma Patients (n=87).
| Clinical feature | n | LINC00313 | p value | |
|---|---|---|---|---|
| High | Low | |||
| Age | 0.761 | |||
| ≥20 yr | 27 | 13 | 14 | |
| <20 yr | 60 | 31 | 29 | |
| Gender | 0.914 | |||
| Man | 47 | 22 | 25 | |
| Woman | 40 | 22 | 18 | |
| Tumor size | 0.012 | |||
| ≥5 cm | 38 | 25 | 13 | |
| <5 cm | 49 | 19 | 30 | |
| Clinical stage | <0.001 | |||
| II A–B | 37 | 9 | 28 | |
| III A–B | 50 | 35 | 15 | |
| Distant metastasis | 0.024 | |||
| Yes | 48 | 30 | 18 | |
| No | 39 | 14 | 25 | |
Chi-square test was used to analyze correlations between LINC00313 expression and clinicopathological parameters in these 87 osteosarcoma patients.
Cells and cell culture
Four human OS cell lines (U2OS, MG-63, Saos-2, and SOSP-9607) and one normal osteoblast cell line [hFOB 1.19 (human fetal osteoblastic cell line)] were purchased from ATCC (Shanghai, China).22 These cells were cultured in DMEM (Gibco, Carlsbad, CA, USA) containing 10% FBS (Gibco) and 1% penicillin/streptomycin at 37℃.
Lentivirus infection and cell transfection
siLINC00313, miR-342-3p mimic/NC, and miR-342-3p inhibitor/NC were purchased from Ribobio (Guangzhou, China). Cell transfection of oligonucleotides into U2OS and MG-63 cells was performed by Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer's instructions.
A lentiviral system (Applied Biological Materials, Ontario, Canada) to stably reduce LINC00313 expression (Lv-shLINC00313) and its negative control vector (Scramble) in the cells were developed following the manufacturer's instructions.
Total RNA isolation and real-time quantitative PCR
Total RNA from tissue samples and cultured cells was isolated using TRIzol reagent (Thermo, Waltham, MA, USA) following the manufacturer's protocol. The first strands of cDNA were synthesized dependent on total RNA using reverse transcription kits (Abcam, Cambridge, UK), and amplification of cDNA was performed using SYBR Premix Ex Taq Master Mix (Invitrogen). Real-time quantitative PCR (qPCR) was conducted on an Applied Biosystem 7500 Real-Time PCR System (Thermo), and the expression of selected mRNAs and miR-342-3p were calculated according to the comparative threshold cycle value (2−ΔΔCt) method, compared with β-actin (for mRNA) or U6 small nuclear RNA (U6, for miRNA). PCR primers for LINC00313, miR-342-3p, U6, and β-actin were purchased from Invitrogen. All operations were conducted at least three times.
Total protein extraction and Western blotting
Total protein was extracted from treated cells with RIPA lysis buffer (Beyotime, Shanghai, China) to measure protein expression, including cleaved caspase 3, FOSL2, and GAPDH. Western blotting was performed according to standard procedures, and GAPDH on the same membrane was used as a loading control. The primary antibodies were as follows: anti-cleaved caspase 3 (#9664, 1:2000, CST; Danvers, MA, USA); anti-FOSL2 (#19967, 1:2000, CST); anti-GAPDH (#9484, 1:5000, Abcam). The proteins were visualized using ECL procedure, and ImageJ (National Institutes of Health, Bethesda, MD, USA) was utilized to analyze the gray intensities of bands.
Cell viability assay (with MTT)
3-(4,5-dimethylthiazole-2-y1)-2,5-biphenyl tetrazolium bromide (MTT) was used to evaluate cell viability and cell proliferation. 5 mg/mL of MTT was added to DMEM, and cells were incubated for 4 h, followed with incubation of 100 µL of DMSO. The spectrophotometric absorbance of each sample was measured at 570 nm. The experiments were conducted at least three times.
Flow cytometry
OS cells with different processing methods were analyzed by flow cytometry. We used Annexin V-FITC/PI kit (Beyotime) to quantify apoptotic cells in accordance with the manufacturer's protocol. Fluorescence was analyzed on a cytoFLEX LX flow cytometer (Beckman-Counter Electronics, Jiangsu, China) using CytExpert software.
Transwell assay for cell migration and invasion
For migration and invasion assay,23,24 U2OS2 and MG-63 cells supplemented with 200 µL of serum-free medium were plated in the upper chamber with a non-coated and Matrigel-coated membrane (Corning Inc., Corning, NY, USA). Medium containing 10% FCS was used as a chemo-attractant and loaded in the low chamber. Transwell systems were stored at 37℃ for 48 h. The invaded and migrated cells into the lower chambers were stained with crystal violet and quantitated under microscopy while taking pictures.
Luciferase report assay
Plasmids of pGL3-basic, pGL3-LINC00313 wt, or pGL3-LINC00313 mut were co-transfected with miR-342-3p mimic/miR-NC mimic or miR-342-3p inhibitor/miR-NC inhibitor into 293T cells; pGL3-basic, pGL3-FOSL2 wt, or pGL3-FOSL2 mut was co-transfected with miR-342-3p mimic or miR-NC mimic into 293T cells. All transfection procedures were performed by Lipofectamine 2000 (Invitrogen). After 48 h-transfection, luciferase activity was measured using a dual-luciferase reporter system (Promega, Madison, WI, USA). The ratio of Firefly to Renilla luciferase activity was used as the relative luciferase activity. All experiments were repeated three times.
RNA immunoprecipitation and RNA pull-down assay
RNA immunoprecipitation and RNA pull-down assays were performed with MG-63 cell extract. Magna RIP™ RNA-binding protein immunoprecipitation kits (MilliporeSigma, Billerica, MA, USA) were utilized to detect expression of LINC00313 from the samples bound to the Ago2 antibody or IgG,25 and LINC00313 levels were examined using qPCR in samples pulled down by biotin-labeled miR-342-3p or NC.26 All experiments were conducted following standard instructions.
Xenograft mouse model
Eight-week-old athymic mice were obtained from the Sixth Affiliated Hospital of Xinjiang Medical University. The animal experiments were approved by the Institutional Review Board of the Sixth Affiliated Hospital of Xinjiang Medical University and were performed in accordance with the National Institutions of Health Guide for Care and Use of Laboratory Animals. In vivo experiments referred to previous research.27 Equal numbers (106) of U2OS and MG-63 cells/0.2 mL with forced expressed siLINC00313 or Scramble were injected subcutaneously in athymic mice (5 mice per group) for 25 days. The tumors were measured with a caliper every 5 days for 5 times till the 25th day, and the mice were euthanized on day 25. Tumor volumes were calculated using the following formula: V (mm3)=1/2ab2, where a is the longest tumor axis and b is the shortest tumor axis. The weight of tumors was evaluated with an electronic scale. Immediately, tumors were frozen in −80℃ for further isolation of total RNA and protein.
Statistical analyses
Data are presented as the mean±standard error. Two-group comparisons were performed using Student's t-test on SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). p<0.05 was considered statistically significant. The relationship between LINC00313 and miR-342-3p expression levels in 87 OS tissue samples was analyzed by Spearman's correlation analysis. Survival analysis for high and low expression of LINC00313 (compared with the median) was conducted using Kaplan-Meier curves. The association between LINC00313 expression and clinicopathological data of this cohort of OS patients was analyzed using the chisquare test.
RESULTS
Roles of LINC00313 in OS tissues and cell lines
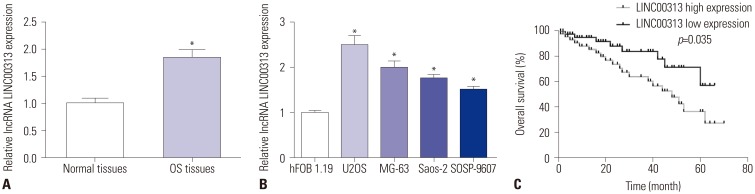
First, we detected the roles of LINC00313 in OS patients. As shown in Fig. 1A, LINC00313 was upregulated in OS tissues (n=87), compared with paired adjacent normal tissues (n=24). This cohort of patients was divided into high and low LINC00313 expression groups according to the median LINC00313 level. With the chi-square test, we discovered that LINC00313 expression was associated with tumor size (≥5 cm), TNM stage, and distant metastasis (Table 1). In vitro, relative expression levels of LINC00313 were also higher in human OS cell lines, including U2OS, MG-63, Saos-2 and SOSP-9607, than in hFOB 1.19 cells (Fig. 1B). In addition, as shown in Fig. 1C, the OS patients were divided into high and low LINC00313 expression groups depending on the median LINC00313 expression level. The 60-month survival rate of high LINC00313 expression was lower than 40%, and that of low LINC00313 expression was 60%. Moreover, over 80% of OS patients with high LINC00313 levels survived less than 20 months. These data indicated that LINC00313 is upregulated in OS tissues and cell lines and that high expression of LINC00313 confers poor prognosis among OS patients.
Fig. 1. Roles of lncRNA LINC00313 in osteosarcoma (OS) tissues and cell lines. (A) Expression of LINC00313 in OS tissues and normal tissues was detected by qPCR. (B) Expression of LINC00313 in OS cells (U2OS, MG-63, Saos-2, and SOSP-9607) and normal osteoblast cells (hFOB 1.19) was detected by qPCR. (C) Survival analysis of differential expression of LINC00313 with Kaplan-Meier curves. All experiments were carried out three times. *p<0.05.
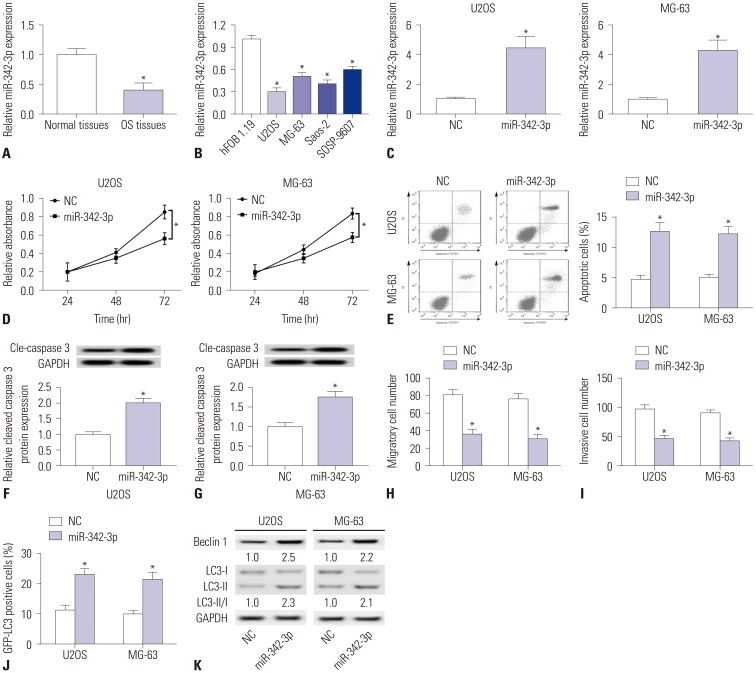
Fig. 3. Roles of miR-342-3p in osteosarcoma (OS) tissues and cell lines. (A) Expression of miR-342-3p in OS tissues and normal tissues was detected by qPCR. (B) Expression of miR-342-3p in OS cells (U2OS, MG-63, Saos-2, and SOSP-9607) and normal osteoblast cells (hFOB 1.19) was detected by qPCR. (C-K) OS cell lines (U2OS and MG-63) were transfected with miR-342-3p mimic (miR-342-3p) and miR-NC mimic (NC) for further study. (C) Expression of miR-342-3p was detected after transfection. (D and E) Cell proliferation and apoptosis were measured by MTT staining and flow cytometry. (F and G) Expression of Cle-caspase 3 was detected with Western blotting. (H and I) Cell migration and invasion were assayed with transwell assay. (J) GFP-LC3 positive cells were recorded after being transfected with GFP-LC3 plasmid. (K) Expressions of Beclin 1, LC3-I, and LC3-II were detected with Western blotting. GAPDH was the loading control. All experiments were carried out three times. *p<0.05.
Effects of LINC00313 knockdown in OS cell lines
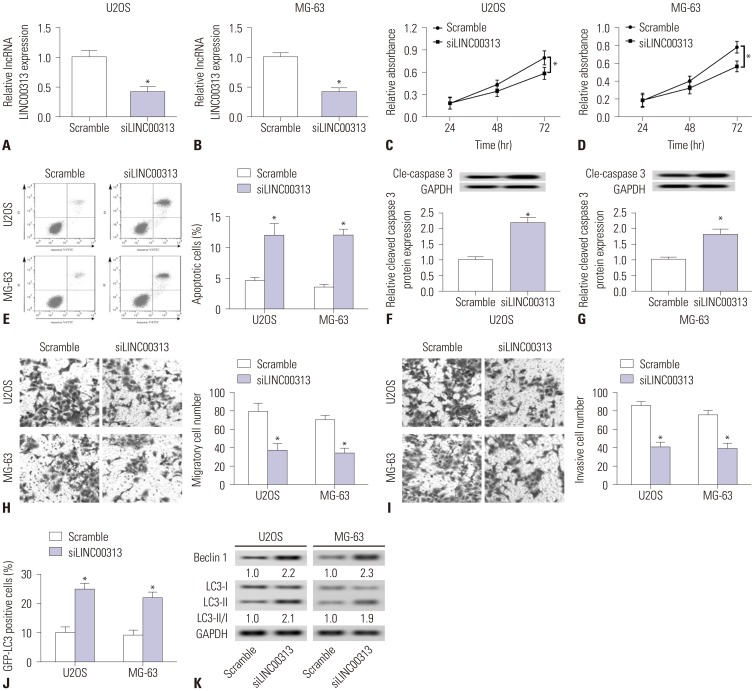
Considering that LINC00313 expression was considerably high in OS cells (Fig. 1B), we transfected siLINC00313 and scrambled siRNA into U2OS and MG-63 cells to study the effects of LINC00313 knockdown. In doing so, we successfully downregulated LINC00313 in cells (Fig. 2A and B). Then, we investigated the cell toxicity of LINC00313 knockdown. There was no significant difference after transfection for 24 h and 48 h in cells (Fig. 2C and D); however, siLINC00313 cells (siLINC00313) showed less cell viability than scrambled siRNA cells at 72 h. In addition, apoptotic cell numbers increased among cells (Fig. 2E). Similarly, expression of cleaved caspase 3 increased (Fig. 2F and G). Next, we investigated the effects of LINC00313 knockdown on tumor cell metastasis. The results revealed fewer migratory and invasive cells (Fig. 2H and I). Last, we studied the effects of LINC00313 knockdown on autophagy. GFP-LC3 plasmid was transfected into U2OS and MG-63 cells, followed by detection of positive cells. Data revealed an increase in GFPLC3 positive cells (Fig. 2J) and expressions levels of Beclin 1 and LC3-II/I (Fig. 2K). These results indicated that LINC00313 knockdown promotes cell apoptosis and autophagy, suppresses cell proliferation, migration, and invasion in OS cells U2OS and MG-63.
Fig. 2. Effects of LINC00313 knockdown in osteosarcoma (OS) cell lines. OS cell lines (U2OS and MG-63) were transfected with siLINC00313/Scramble for further study. (A and B) Expression of LINC00313 was detected by qPCR. (C–E) Cell proliferation and apoptosis were measured using MTT staining and flow cytometry. (F and G) Expression of Cle-caspase 3 was detected with Western blotting. (H and I) Cell migration and invasion were assayed with transwell assay (magnification, ×100). (J) GFP-LC3 positive cells were recorded after being transfected with GFP-LC3 plasmid. (K) Expressions of autophagyrelated proteins (Beclin 1, LC3-I, and LC3-II) were detected with Western blotting. GAPDH was the loading control. All experiments were carried out three times. *p<0.05.
Roles of miR-342-3p in OS tissue and cell lines
To investigate the lncRNA-miRNA-mRNA pathway underlying LINC00313 in OS, we first searched for potential miRNAs targeting LINC00313. According to the in cilico data on starBase v.2.0 (http://starbase2/browse/LINC00313), there were a total of 13 miRNAs possessing the complementary binding sites to LINC00313. Among these miRNAs, four miRNAs, including miR-199a/b-5p and miR-19a/b-3p, were upregulated, and nine miRNAs were downregulated in this cohort of OS patients (n=87), compared to 24 adjacent normal tissues (data not shown). The regulatory effect of LINC00313 silencing on miR-342-3p was most effective, as reflected in the highest levels of miR-342-3p (Supplementary Fig. 1, only online). Thus, miR-342-3p was the preferred miRNA in this study.
We detected the roles of miR-342-3p in OS tissues and cells. As shown in Fig. 3A, miR-342-3p was downregulated in OS tissues, and relative expression levels of miR-342-3p were lower in U2OS, MG-63, Saos-2, and SOSP-9607 cells (Fig. 3B). We detected miR-342-3p expression levels to ensure the overexpression of miR-342-3p in U2OS and MG-63 cells (Fig. 3C). We originally investigated whether miR-342-3p overexpression elicited cell toxicity. MiR-342-3p mimic cells showed increased cell viability after transfection for 72 h, both in U2OS and MG-63 cells (Fig. 3D). Along with statistical increases in apoptotic cell numbers (Fig. 3E), similar results were obtained for the expression of cleaved caspase 3 (Fig. 3F and G). Second, we investigated the effects of miR-342-3p overexpression on tumor cell metastasis and observed decreased migratory and invasive cells (U2OS and MG-63) (Fig. 3H and I). Later, we studied the effects of miR-342-3p overexpression on autophagy. The results revealed an increase in GFP-LC3 positive cells (Fig. 3J) and expression levels of Beclin 1 and LC3-II/I (Fig. 3K) in U2OS and MG-63 cells. This suggests that miR-342-3p overexpression promotes cell apoptosis and autophagy and suppresses cell proliferation and cell migration and invasion in OS cell lines (U2OS and MG-63), consistent with LINC00313 knockdown.
LINC00313 regulates miR-342-3p by targeted binding
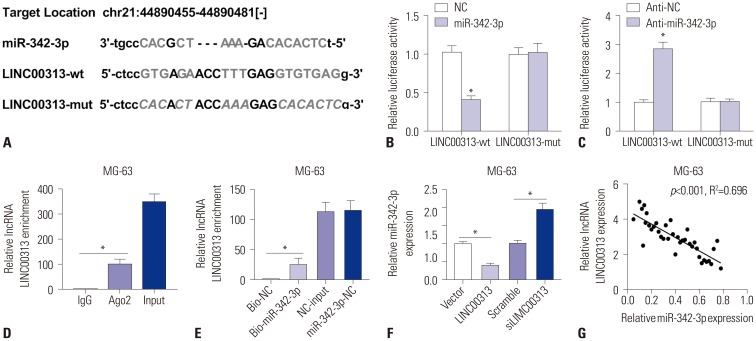
A possible target of LINC00313 was retrieved and identified as miR-342-3p on starBase v.2.0 (Fig. 4A). The sequences of the putative LINC00313 binding sites were shown in wild-type and mutant LINC00313-3′ UTR. To confirm this, a dual-luciferase reporter assay was performed. Fig. 4B and C showed that relative luciferase activity decreased in 293T cells co-transfected with LINC00313-wt and miR-342-3p mimic, and elevated in 293T cells co-transfected with LINC00313-wt and anti-miR-342-3p. We examined the enrichment of LINC00313 from Ago2 immunoprecipitation (Fig. 4D) and bio-miR-342-3p mimic (Fig. 4E) Furthermore, miR-342-3p levels in MG-63 cells (Fig. 4F) were found to be upregulated upon transfection with siLINC00313 and to be downregulated after transfection with LINC00313 overexpression plasmid. These results suggested that LINC00313 directly inhibits miR-342-3p expression through a specific binding site located in 3′ UTR. Moreover, a significant inverse correlation between LINC00313 and miR-342-3p was also observed in 37 OS tissue samples (Fig. 4G). Taken together, we deemed that LINC00313 negatively regulates miR-342-3p by targeted binding in a linearly dependent manner.
Fig. 4. LINC00313 regulates miR-342-3p by targeted binding. (A) Prediction of the potential binding sites between LINC00313 and miR-342-3p on starBase v.2.0. (B and C) Luciferase report assay was performed to determine the luciferase activity of 293T cells transfected between either miR-342-3p/NC or anti-miR-342-3p/anti-NC and LINC00313-wt/LINC00313-mut. (D) RNA immunoprecipitation was conducted to determine LINC00313 expression in MG-63 cell samples. (E) Determination of LINC00313 mRNA in MG-63 cells after transfected with Bio-miR-342-3p/Bio-NC using qPCR. (F) Expression of miR-342-3p in MG-63 cells after being transfected with LINC00313 overexpression plasmid/empty plasmid (LINC00313/Vector), siLINC00313/scrambled siRNA (Scramble), separately. (G) Spearman's correlation analysis between miR-342-3p expression and LINC00313 mRNA levels in 37 OS tissue samples. All experiments were carried out in triplicate. *p<0.05.
Effects of restoration of miR-342-3p in OS cell lines
Next, we wondered whether miR-342-3p restoration could ameliorate OS characteristics. Restoration of miR-342-3p in U2OS and MG-63 cells was induced by co-transfection with siLINC00313 and anti-NC, and defective miR-342-3p restoration was obtained by incubation of siLINC00313 and anti-miR-342-3p. First and foremost, we verified high expression of siLINC00313-induced miR-342-3p, as shown in Fig. 5A. We noted the following differences between high siLINC00313-induced miR-342-3p expression group and control group: reduced cell proliferation (Fig. 5B), increased apoptosis (Fig. 5C–E), increased cleaved caspase 3 protein levels (Fig. 5D and E), fewer migratory (Fig. 5F) and invasive (Fig. 5G) cells, and the promotion of GFP-LC3 positive cells (Fig. 5H) and Beclin 1 and LC3-II/I expression (Fig. 5I). All of these results indicated that high siLINC00313-induced miR-342-3p expression exerts anti-tumor effects in U2OS and MG-63 cells, in agreement with that of high miR-342-3p mimic-induced expression (Fig. 3D–K).
Fig. 5. Effects of restoration of miR-342-3p in osteosarcoma (OS) cell lines. Restoration of miR-342-3p in U2OS and MG-63 cells was induced by co-transfection with siLINC00313 and anti-NC, and defective miR-342-3p restoration was obtained by incubation of siLINC00313 and anti-miR-342-3p. (A) Expression levels of miR-342-3p were detected by qPCR. (B) Cell proliferation was assessed by MTT staining. (C) Cell apoptosis was detected with flow cytometry. (D and E) Activity of caspase 3 was assessed using Western blotting. (F and G) Cell migration and invasion were determined using transwell assay. (H) Positive cells of GFP-LC3 were calculated after transfection. (I) Expressions of Beclin 1, LC3-I, and LC3-II were detected with Western blotting. GAPDH was the loading control. All experiments were carried out in triplicate. *p<0.05.
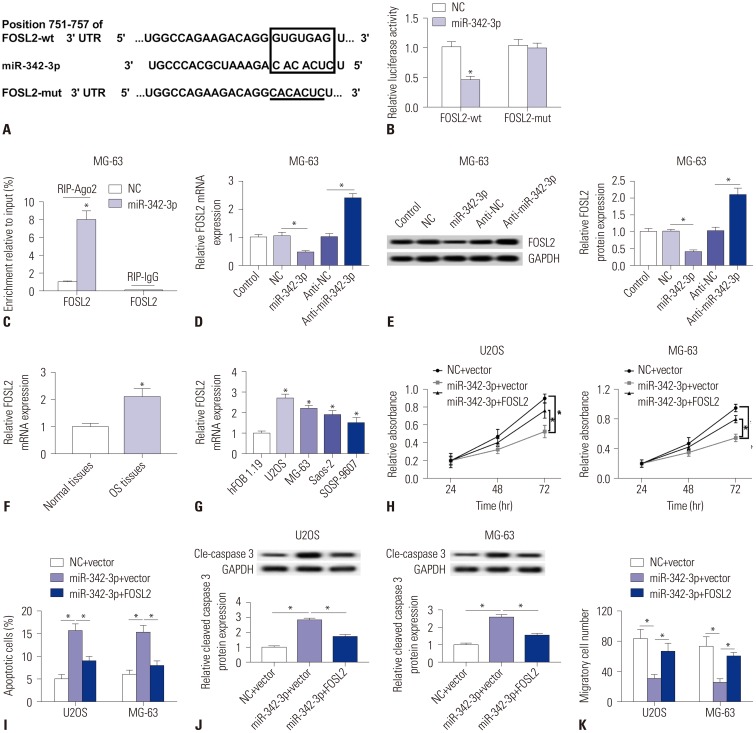
MiR-342-3p modulates OS characteristics by targeting FOSL2
Previous study claimed that FOSL2 was upregulated in OS tissues and cell lines,21 we used the TargetScan Human to retrieve and identified FOSL2 as a possible target gene for miR-342-3p (Fig. 6A). The sequences of the putative miR-342-3p binding sites (GUGUGAG) were on position 751–757 of FOSL2 3′ UTR, and which were mutated into CACACUC. To confirm this, a dual-luciferase report assay was performed. And, Fig. 6B told us that relative luciferase activity dropped in 293T cells co-transfected with FOSL2-wt and miR-342-3p mimic. Further, the outcome (Fig. 6C) was that FOSL2 was dramatically enriched from Ago2. Furthermore, FOSL2 was found to be downregulated (Fig. 6D and E) after transfected with miR-342-3p mimic, and upregulated (Fig. 6D and E) after transfected with anti-miR-342-3p. Above results suggested that miR-342-3p directly inhibited FOSL2 expressions in MG-63 cells through specific binding site located in 3′ UTR. Moreover, FOSL2 was highly expressed in both OS tissues and cell lines (Fig. 6F and 6G). Subsequent experiments exhibited big differences between miR-342-3p mimic-induced FOSL2 lower expression group and control group: declined cell proliferation (Fig. 6H), increased apoptosis (Fig. 6I) and raised cleaved caspase 3 protein levels (Fig. 6J); reduction of migratory (Fig. 6K) and invasive (Fig. 6L) cells; promoted GFP-LC3 positive cells (Fig. 6M) and expression of Beclin 1 and LC3-II/I (Fig. 6N). All these results indicated miR-342-3p mimic-induced FOSL2 lower expression played an antitumor role in U2OS and MG-63 cells, in agreement with that of miR-342-3p high expression (Figs. 3 and 5) and LINC00313 knockdown (Fig. 1). Furthermore, exogenous administration of FOSL2 overexpression vector could partially block the effect of miR-342-3p upregulation in U2OS and MG-63 cells (Fig. 6H–6N).
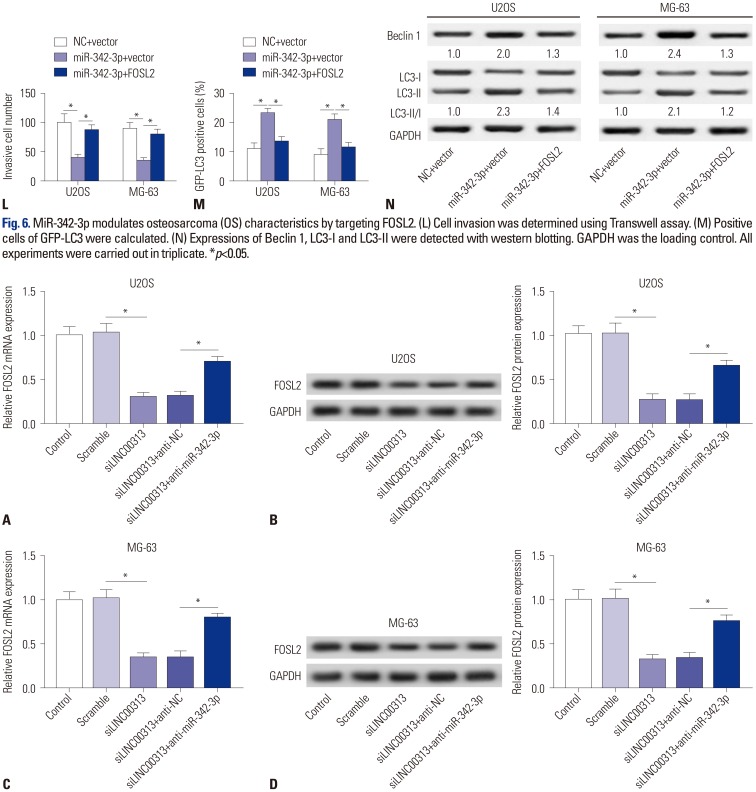
Fig. 6. MiR-342-3p modulates osteosarcoma (OS) characteristics by targeting FOSL2. (A) Prediction of potential binding sites between miR-342-3p and FOSL2 on Targetscan Human. (B) Luciferase reporter assay was performed to determine the luciferase activity of 293T cells transfected with miR-342-3p mimics/miR-NC mimic (miR-342-3p/NC) and FOSL2-wt/FOSL2-mut. (C) RNA immunoprecipitation was conducted to determine FOSL2 expressions in MG-63 cell samples. (D and E) Determination of FOSL2 expression both at the mRNA level and protein level in MG-63 cells after transfection with miR-342-3p/NC, anti-miR-342-3p/anti-NC. Restoration of FOSL2 in U2OS and MG-63 cells was induced by co-transfection with miR-342-3p mimic and empty plasmid (Vector), and defective FOSL2 restoration was obtained by incubation of miR-342-3p mimic and FOSL2 overexpression plasmid (FOSL2). (F and G) qPCR detected FOSL2 mRNA expression in OS tissues and cell lines. (H) Cell proliferation was assessed by cell activity with MTT staining. (I) Cell apoptosis was detected with flow cytometry. (J) Activity of caspase 3 was measured using western blotting. (K) Cell migration was determined using Transwell assay. GAPDH was the loading control. (L) Cell invasion was determined using Transwell assay. (M) Positive cells of GFP-LC3 were calculated. (N) Expressions of Beclin 1, LC3-I and LC3-II were detected with western blotting. GAPDH was the loading control. All experiments were carried out in triplicate. *p<0.05.
LINC00313 positively regulates FOSL2 by sponging miR-342-3p
Further experiments on whether LINC00313 functions in OS through a LINC00313/miR-342-3p/FOSL2 axis showed that FOSL2 was downregulated by siLINC00313 at both the mRNA level and protein level in U2OS cells (Fig. 7A and B), whereas FOSL2 expression was further rescued upon co-incubation with siLINC00313 and anti-miR-342-3p, compared with siLINC00313 and anti-NC. Importantly, similar results were acquired in MG-63 cells (Fig. 7C and D). We came to the conclusion that LINC00313 knockdown inversely regulates FOSL2 expression, which could be abolished reducing miR-342-3p in OS cell lines.
Fig. 7. LINC00313 positively regulates FOSL2 by sponging miR-342-3p. Effects of LINC00313 on FOSL2 expression in U2OS and MG-63 cells were illuminated by transfection with siLINC00313/Scramble or co-transfection of siLINC00313 and anti-miR-342-3p/anti-NC. FOSL2 expression was determined using qPCR and Western blotting in U2OS (A and B) and MG-63 (C and D) cells. All experiments were carried out in triplicate. *p<0.05.
Effects of LINC00313 knockdown on tumor growth
Finding that LINC00313 promotes tumor characteristics in OS cell lines through miR-342-3p-mediated FOSL2 upregulation, we then wondered whether LINC00313 knockdown could suppress tumor growth in vivo. Lv-ShLINC00313 was infected intoU2OS and MG-63 cells, which were implanted in subcutaneous areas of athymic mouse for 25 days. As Fig. 8A and B show, tumor volumes greatly reduced after implantation for 15 days, as did tumor weight. After 25 days, LINC00313 and FOSL2 expression was downregulated (Fig. 8C, D, G, H), while miR-342-3p was upregulated (Fig. 8E and F) in tumors in athymic mouse. All data supported the idea that LINC00313 knockdown inhibits OS growth in vivo.
Fig. 8. Effects of LINC00313 knockdown on tumor growth. Lv-ShLINC00313 was infected into U2OS and MG-63 cells, which were implanted in subcutaneous areas of athymic mouse for 25 days. (A and B) Measurement of tumor volume every 5 days, and the weights of tumors were determined on day 25, respectively. (C and D) Detection of LINC00313 expression levels in tumors on day 25. (E and F) Expression levels of miR-342-3p in tumors were measured on day 25. (G and H) Expression levels of FOSL2 protein in tumors were measured on day 25. All experiments were carried out in triplicate. *p<0.05.
DISCUSSION
Here, we report the role of LINC00313 in OS, which was upregulated in OS tissues and cells. High expression of LINC00313 was associated with tumor size, TNM stage, and distant metastasis, as well as a shorter overall survival rate. Meanwhile, LINC00313 knockdown conferred good prognosis presumably through a miR-342-3p/FOSL2 axis.
LINC00313 has been shown to be upregulated and correlated with a poor prognosis in cancers. LINC00313 is a novel lncRNA reported to be highly expressed in both T2- and N1-stage LC and to indicate shorter overall survival, which suggests that LINC00313 could be a biomarker of LC.11 In the study, we found that the 60-month survival rate of individuals with high LINC00313 expression was about 40%, while that of individuals with low LINC00313 expression was over 60%. Our findings are in line with a very recent study that noted a strong association of LINC00313 with lymph node metastasis, advanced tumor stage, and TNM stage in PTC patients.12 These data support that LINC00313 could be a potential biomarker for diagnosing cancers, including OS, PTC, and LC.
LINC00313 functions as oncogene in cancers via sponging miRNAs. Although studies of LINC00313 are scarce, research has revealed that LINC00313 plays a cancer-promotion role in PTC,12 which agrees with the effect of LINC00313 in OS reflected in our results: Downregulation of LINC00313 inhibited the proliferative, migratory, and colony-forming abilities of PTC cells; silencing LINC00313 induced cell cycle arrest and apoptosis in PTC cells.12 In our study, LINC00313 knockdown promoted cell apoptosis and autophagy and suppressed cell proliferation, migration, and invasion in OS cell lines (U2OS and MG-63), leading to reduced tumor growth in a xenograft mouse model. Functional experiments showed overexpression of miR-4429 could abrogate the oncogenic role of LINC00313 in PTC cells, implying that LINC00313 is biologically active via sponging miR-4429. Similarly, LINC00313 suppressed OS tumor characteristics through miR-342-3p.
MiR-342-3p inhibits cell proliferation, migration, and invasion in cancers. Here, we discerned that downregulation of miR-342-3p occurs in OS tissues and cells and that overexpression of miR-342-3p elevated apoptosis and autophagy and reduced cell viability, migration and invasion. Moreover, high miR-342-3p expression promoted OS development by directly downregulating FOSL2. Other researchers have proposed that AEG-1 is a target gene of miR-342-3p,16 which we found to inhibit proliferation, migration, and invasion of OS and other have found to participate in NF-κB signaling pathway.28 High expression of miR-4429 has been found to result in a loss of Bcl-2 and Mcl-2, allowing Bax2 to mediate LINC00313 silencing-induced apoptosis.12 In this study, LINC00313 knockdown triggered cell autophagy and apoptosis, at least, depending on caspase cascade.
FOSL2 has been identified as a target of several miRNAs to promote cancer cell metastasis. As discussed in the present study, FOSL2 was downregulated, and miRNAs for several antioncogenes inhibited FOSL2 expression by target binding. FOSL2 downregulation appears to be a critical step in regulation of OS and hepatocellular carcinoma properties by miR-143-3p and miR-133a through TGFβ/Smad signaling pathway: 20,21 FOSL2 overexpression in normal colonic epithelial cells (FHC cells) induced pro-mesenchymal cell features, and FOSL2 silencing inhibited cell migration and invasion ability in LoVo cells.18 MiR-30e enhanced hepatocyte proliferation and decreased hepatocyte apoptosis in septic rats.29 MiR-597 suppressed cell proliferation, migration, and invasion in breast cancer by directly upregulating FOSL2.19
The potential role and clinical value of LINC00313 in human cancers remain elusive, and a further research of LINC00313 is warranted. For one, the molecular mechanism of how LINC00313 functions in OS needs to be to be uncovered, especially in regards to signaling pathways, such as TGFβ/Smad. Additionally, the biological functions of LINC00313 need to be explored among diseases, including tumors and cancers. Additionally, gene regulatory networks related with LINC00313 and other factors await discovery.
The median survival time of OS patients is only 23 months. Current clinical trials have failed to improve OS survival time, and there is in urgent need for deeper knowledge of the occurrence, development, and prognosis of OS. We identified for the first time an anti-tumor role for LINC00313 in suppressing cell proliferation, migration, and invasion, and promoting cell apoptosis and autophagy through an miR-342-3p/FOSL2 axis. Our results provide new insights into OS and suggest that knockdown of LINC00313, a novel lncRNA, and/or overexpression of miR-342-3p could serve as a new approach for OS treatment in clinical trials.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Hongtao Chen.
- Data curation: Hongtao Chen, Paerhati Wahafu, Leilei Wang.
- Formal analysis: Hongtao Chen.
- Investigation: Hongtao Chen, Leilei Wang.
- Methodology: Hongtao Chen.
- Resources: Paerhati Wahafu, Xuan Chen.
- Software: Paerhati Wahafu, Xuan Chen.
- Supervision: Hongtao Chen.
- Validation: Hongtao Chen.
- Visualization: Leilei Wang, Xuan Chen.
- Writing—original draft: Hongtao Chen.
- Writing—review & editing: Xuan Chen.
- Approval of final manuscript: all authors.
SUPPLEMENTARY MATERIAL
The regulatory effect of LINC00313 silencing on miRNAs in U2OS cells. Expression levels of miRNAs were measured by qPCR in U2OS cells transfected with scrambled siRNA (scramble) or siLINC00313. miR-342-3p was the most upregulated miRNA by LINC00313 silencing.
References
- 1.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 2.Slade AD, Warneke CL, Hughes DP, Lally PA, Lally KP, Hayes-Jordan AA, et al. Effect of concurrent metastatic disease on survival in children and adolescents undergoing lung resection for metastatic osteosarcoma. J Pediatr Surg. 2015;50:157–160. doi: 10.1016/j.jpedsurg.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Yu FX, Hu WJ, He B, Zheng YH, Zhang QY, Chen L. Bone marrow mesenchymal stem cells promote osteosarcoma cell proliferation and invasion. World J Surg Oncol. 2015;13:52. doi: 10.1186/s12957-015-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontanella R, Pelagalli A, Nardelli A, D’Alterio C, Ieranò C, Cerchia L, et al. A novel antagonist of CXCR4 prevents bone marrow-derived mesenchymal stem cell-mediated osteosarcoma and hepatocellular carcinoma cell migration and invasion. Cancer Lett. 2016;370:100–107. doi: 10.1016/j.canlet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Pietrovito L, Leo A, Gori V, Lulli M, Parri M, Becherucci V, et al. Bone marrow-derived mesenchymal stem cells promote invasiveness and transendothelial migration of osteosarcoma cells via a mesenchymal to amoeboid transition. Mol Oncol. 2018;12:659–676. doi: 10.1002/1878-0261.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. doi: 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 7.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Long non-coding RNAs in osteosarcoma. Oncotarget. 2017;8:20462–20475. doi: 10.18632/oncotarget.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kun-Peng Z, Chun-Lin Z, Xiao-Long M. Antisense lncRNA FOXF1-AS1 promotes migration and invasion of osteosarcoma cells through the FOXF1/MMP-2/-9 pathway. Int J Biol Sci. 2017;13:1180–1191. doi: 10.7150/ijbs.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiscon G, Conte F, Farina L, Paci P. Network-based approaches to explore complex biological systems towards network medicine. Genes (Basel) 2018;9:E437. doi: 10.3390/genes9090437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Qiu M, Xu Y, Mao Q, Wang J, Dong G, et al. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol. 2015;36:9969–9978. doi: 10.1007/s13277-015-3714-6. [DOI] [PubMed] [Google Scholar]
- 12.Wu WJ, Yin H, Hu JJ, Wei XZ. Long noncoding RNA LINC00313 modulates papillary thyroid cancer tumorigenesis via sponging miR-4429. Neoplasma. 2018;65:933–942. doi: 10.4149/neo_2018_180219N125. [DOI] [PubMed] [Google Scholar]
- 13.Assmann TS, Recamonde-Mendoza M, de Souza BM, Bauer AC, Crispim D. MicroRNAs and diabetic kidney disease: systematic review and bioinformatic analysis. Mol Cell Endocrinol. 2018;477:90–102. doi: 10.1016/j.mce.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Ranjha R, Aggarwal S, Bopanna S, Ahuja V, Paul J. Site-specific microRNA expression may lead to different subtypes in ulcerative colitis. PLoS One. 2015;10:e0142869. doi: 10.1371/journal.pone.0142869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunz M. MicroRNAs in melanoma biology. Adv Exp Med Biol. 2013;774:103–120. doi: 10.1007/978-94-007-5590-1_6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Liu L, Lv Z, Li Q, Gong W, Wu H. MicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1) Oncol Res. 2017;25:1505–1515. doi: 10.3727/096504017X14886485417426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies JS, Klein DC, Carter DA. Selective genomic targeting by FRA-2/FOSL2 transcription factor: regulation of the Rgs4 gene is mediated by a variant activator protein 1 (AP-1) promoter sequence/CREB-binding protein (CBP) mechanism. J Biol Chem. 2011;286:15227–15239. doi: 10.1074/jbc.M110.201996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Fang XD, Wang XY, Fei BY. Fos-like antigen 2 (FOSL2) promotes metastasis in colon cancer. Exp Cell Res. 2018;373:57–61. doi: 10.1016/j.yexcr.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 19.He J, Mai J, Li Y, Chen L, Xu H, Zhu X, et al. miR-597 inhibits breast cancer cell proliferation, migration and invasion through FOSL2. Oncol Rep. 2017;37:2672–2678. doi: 10.3892/or.2017.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, Guo Z, Sun J, Li J, Dong Z, Zhang Y, et al. MiR-133a acts as an anti-oncogene in hepatocellular carcinoma by inhibiting FOSL2 through TGF-β/Smad3 signaling pathway. Biomed Pharmacother. 2018;107:168–176. doi: 10.1016/j.biopha.2018.07.151. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Dai G, Yu L, Hu Q, Chen J, Guo W. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep. 2018;8:606. doi: 10.1038/s41598-017-18739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan L, Wu X, Yin X, Du F, Liu Y, Ding X. LncRNA CCAT2 promoted osteosarcoma cell proliferation and invasion. J Cell Mol Med. 2018;22:2592–2599. doi: 10.1111/jcmm.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. 2016;20:4445–4451. [PubMed] [Google Scholar]
- 24.Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24:46. doi: 10.1186/s12929-017-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain R, Devine T, George AD, Chittur SV, Baroni TE, Penalva LO, et al. RIP-Chip analysis: RNA-binding protein immunoprecipitationmicroarray (chip) profiling. Methods Mol Biol. 2011;703:247–263. doi: 10.1007/978-1-59745-248-9_17. [DOI] [PubMed] [Google Scholar]
- 26.Panda AC, Martindale JL, Gorospe M. Affinity pulldown of biotinylated RNA for detection of protein-RNA complexes. Bio Protoc. 2016;6:e2062. doi: 10.21769/BioProtoc.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed Pharmacother. 2018;97:1645–1653. doi: 10.1016/j.biopha.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-κB pathway. Biochem Biophys Res Commun. 2015;457:370–377. doi: 10.1016/j.bbrc.2014.12.119. [DOI] [PubMed] [Google Scholar]
- 29.Ling L, Zhang SH, Zhi LD, Li H, Wen QK, Li G, et al. MicroRNA-30e promotes hepatocyte proliferation and inhibits apoptosis in cecal ligation and puncture-induced sepsis through the JAK/STAT signaling pathway by binding to FOSL2. Biomed Pharmacother. 2018;104:411–419. doi: 10.1016/j.biopha.2018.05.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The regulatory effect of LINC00313 silencing on miRNAs in U2OS cells. Expression levels of miRNAs were measured by qPCR in U2OS cells transfected with scrambled siRNA (scramble) or siLINC00313. miR-342-3p was the most upregulated miRNA by LINC00313 silencing.