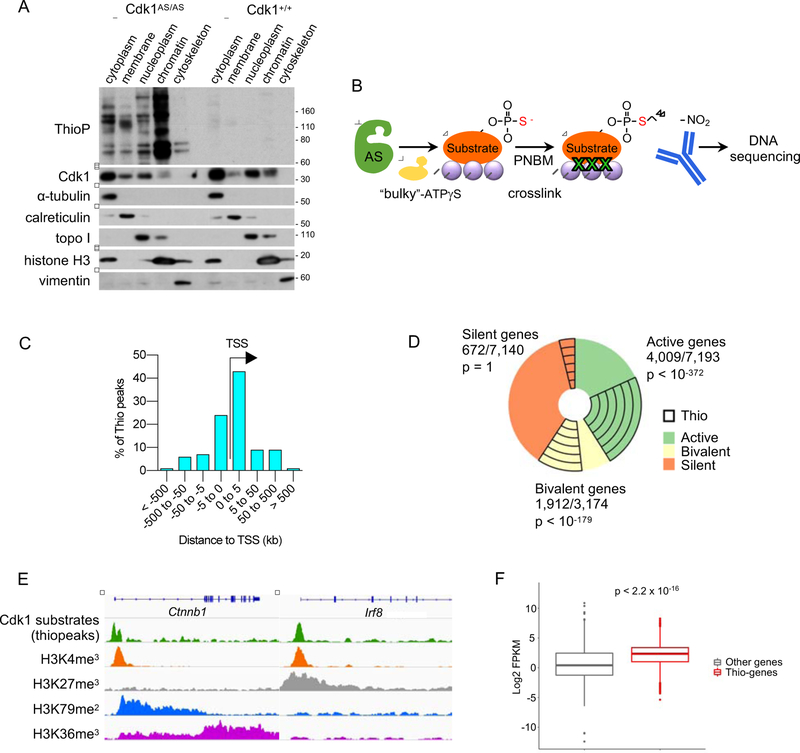
Figure 2. Localization of Cdk1 Substrates on Chromatin.
(A) In vitro cultured Cdk1AS/AS or Cdk1+/+ ES cells were supplemented with 6-Fu-ATP-γ-S for 20 min to thiolabel direct Cdk1 substrates. The indicated fractions were prepared, and analyzed by western blotting with an anti-thiophosphate ester antibody (to detect thio-phosphorylated proteins), or with an anti-Cdk1 antibody. Immunoblots were also probed with antibodies against α-tubulin (marker of the cytoplasmic fraction), calreticulin (membranes), topoisomerase I (topo I, soluble nuclear and chromatin), histone H3 (chromatin) and vimentin (cytoskeleton).
(B) The outline of ‘thio-ChIP-sequencing’ approach. PNBM, p-nitrobenzyl mesylate used to alkylate proteins in order to generate epitopes for an anti-thiophosphate ester antibody.
(C) Distribution of thiopeaks relative to transcription start sites (TSS) of annotated genes.
(D) A pie-chart depicting the fraction of genes with thiopeaks among active, bivalent and silent (containing no H3K4me3 and no H3K79me2 marks) genes.
(E) Examples of thio-ChIP-seq peaks (representing DNA-bound Cdk1 substrates) and histone modification profiles for actively transcribed (Ctnnb1) and bivalent (Irf8) genes. Shown are genome browser tracks for thio-ChIP-seq as well as publicly available ChIP-seq data for the indicated histone marks in mouse ES cells.
(F) Boxplots illustrating expression levels of ES cell genes containing thiopeaks in their promoter regions (Thio-genes) versus of all other genes (Other genes). p < 2.2 × 10−16 (Mann-Whitney test).
See also Table S1.