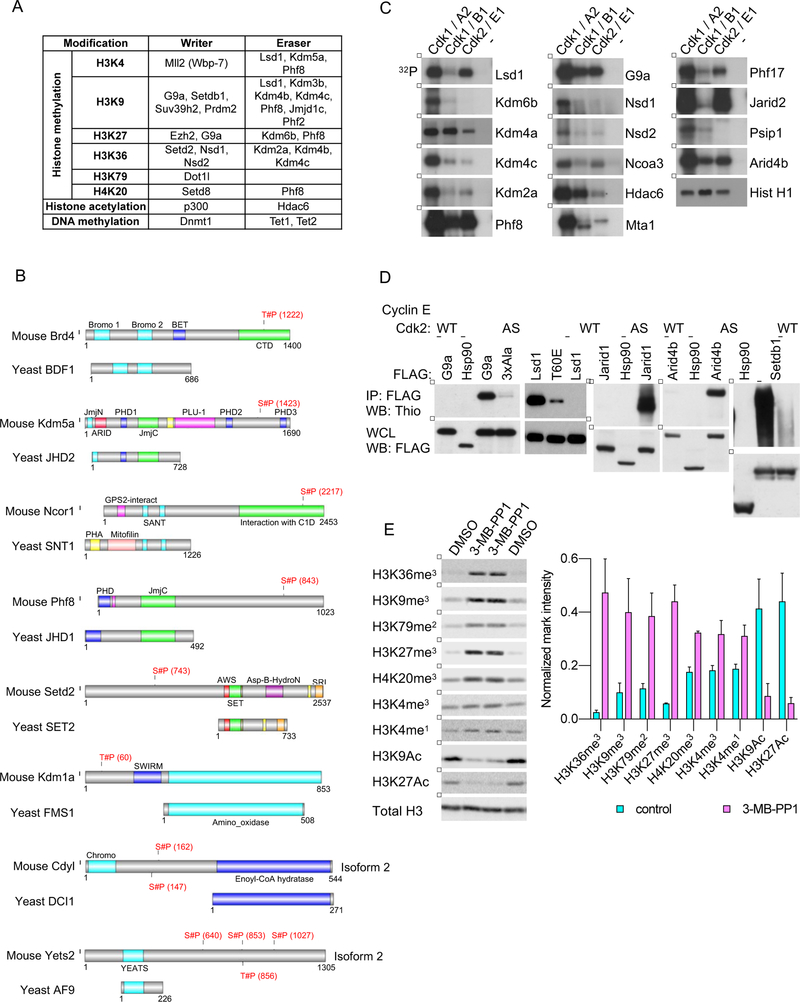
Figure 4. Phosphorylation of Epigenetic Regulators by Cdk1.
(A) A table listing chromatin modifying enzymes (writers and erasers) identified by us as Cdk1 substrates using the direct labeling and/or inhibition approaches.
(B) Examples of murine epigenetic regulators identified by us as Cdk1 substrates, which are phosphorylated by Cdk1 within the domain that is absent in the corresponding ortholog from S. cerevisiae. Shown is alignment of the murine and the corresponding S. cerevisiae protein. Residues of mouse proteins found to be phosphorylated by Cdk1 in inhibition and/or labeling approaches are marked in red.
(C) In vitro kinase reactions with the indicated wild-type cyclin-Cdk kinases, using the indicated epigenetic regulators as substrates. In vitro kinase reactions with the canonical Cdk1 and Cdk2 substrate, histone H1, were used to ensure the comparable specific activity of all cyclin-Cdk kinases used (Hist H1, lowest panel). Lanes marked ‘−’ indicate incubation of substrates with [γ32P]-ATP without any added kinase.
(D) 293T cells were transfected with constructs encoding cyclin E, analog-sensitive Cdk2 (AS), and Flag-tagged versions of the indicated epigenetic regulators. Cells were permeabilized with digitonin and supplemented with N6-furfuryl-ATPγS. Subsequently, epigenetic regulators were immunoprecipitated with an anti-Flag antibody, and immunoblots probed with an anti-thiophosphate ester antibody. As a negative control, Flag-tagged version of Hsp90 (not phosphorylated by cyclin E-Cdk2) was expressed in cells together with cyclin E and analog-sensitive Cdk2. For another negative control, epigenetic regulators were expressed in cells with cyclin E and wild-type Cdk2 (WT). Lower panel, whole cell lysates (WCL) were probed with an anti-Flag antibody. 3xAla and T60E, mutants of G9a and Lsd1, respectively.
(E) Cdk1AS/AS ES cells were left untreated (lanes 1 and 4), or treated with 3-MB-PP1 for 3 (lane 2) or 4 days (lane 3). Chromatin fractions were probed with antibodies against the indicated histone modifications, and against histone H3 (Total H3, loading control). Right, densitometric quantification of the indicated marks, normalized against total histone H3 levels. Mean values ±SD of 2 replicates.
See also Figures S4, S5, Table S4.