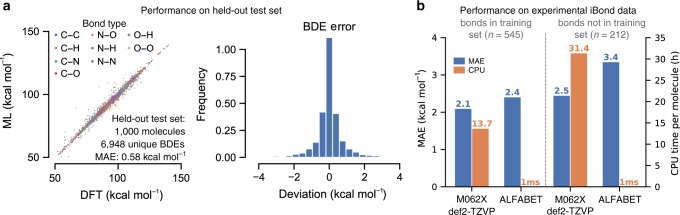
Fig. 3. Performance of the ML BDE prediction algorithm.
a Performance on the held-out, DFT-generated test set. (left) Parity plot of ALFABET predictions vs DFT calculations. BDE points are colored by their bond type. (right) Histogram of prediction errors. The model achieves an MAE of 0.58 kcal mol−1 relative to DFT-calculated values of unseen molecules. b Performance of the model on experimentally measured BDEs from the iBond database. Prediction accuracy was quantified separately for bonds inside the training database (left) and those outside it (right). Molecules and bonds outside the training set tended to be much larger, thus resulting in larger DFT error and long DFT computational times.