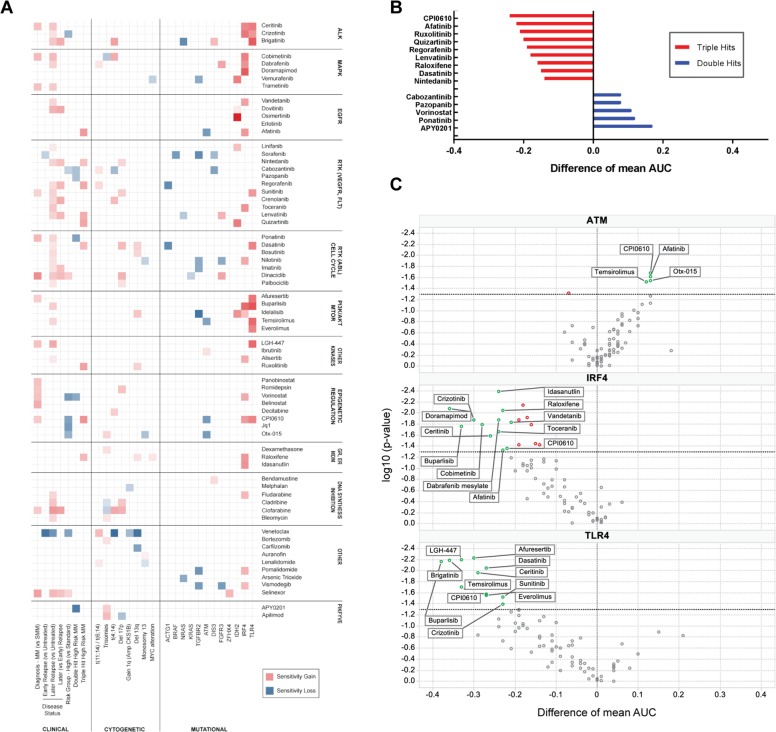
Fig. 4. Clinical stratification, genetic aberrations, and mutational status modulate sensitivity to MMDP agents in MM.
a Differential sensitivity heatmap of the statistically significant associations between drugs and clinical stratification, chromosomal aberration status, and mutational profiles. Red indicates gain of sensitivity in the class under consideration (versus reference or absence of aberration), while blue indicates a loss of sensitivity. The more intense the color, the larger the sensitivity difference. Data are reported for all differential activities with statistical significance (p < 0.05); numerical data and individual box plots are provided in Supplemental Table 9. Signal transduction inhibitors targeting ALK, MAPK, PI3K/AKT, CDKs, and RTKs, as well as HDAC inhibitors and selinexor, had enhanced sensitivity in MM as compared to SMM. KIs across all classes, DNA synthesis inhibitors, and selinexor were more sensitive in later relapse MM samples while venetoclax was more sensitive in untreated samples. Venetoclax, crizotinib, cabozantinib, vorinostat, and bromodomain inhibitors were less responsive in high-risk MM than in standard risk, while selinexor and dinaciclib had increased sensitivity for high-risk samples. b Histogram of differential drug sensitivity (difference of mean AUC between present/absent classes) for double hit high-risk samples (blue) and triple hit high-risk samples (red). Positive values indicate a lower sensitivity while negative values reflect a higher sensitivity in the hit class as compared to high risk (no hit). Double hit high risk is generally associated with drug resistance while triple hit high risk favors drug sensitivity. c Volcano plot of the differential drug sensitivity (difference of mean AUC between present/absent classes) for ATM, IRF4, and TLR4 genes. Positive values indicate a lower sensitivity, while negative values reflect a higher sensitivity in the mutated class as compared to non-mutated class. Drugs with p < 0.05 are labeled on the plot. Mutations in ATM were overall detrimental to sensitivity while mutations in IRF4, and TLR4 were overall favorable to sensitivity. A loss of sensitivity of BET inhibitors CPI0610 and otx-015 was observed in presence of mutations of DNA damage repair associated ATM gene. Inversely, mutations of IRF4 and TLR4 genes associated with a broad increase in sensitivity across multiple drug classes involving signal transduction through receptor and non-receptor kinases. Indeed, samples harboring IRF4 mutations, which have previously been linked to better prognosis were exquisitely sensitive to ALK inhibitors ceritinib and crizotinib as well as 3 out of the 5 tested MAPK inhibitors (cobimetinib, dabrafenib, and doramapimod). When compared to non-mutated samples, samples with TLR4 mutations had better sensitivity to all three tested ALK inhibitors as well as 4 out of 5 PI3K/mTOR inhibitors.