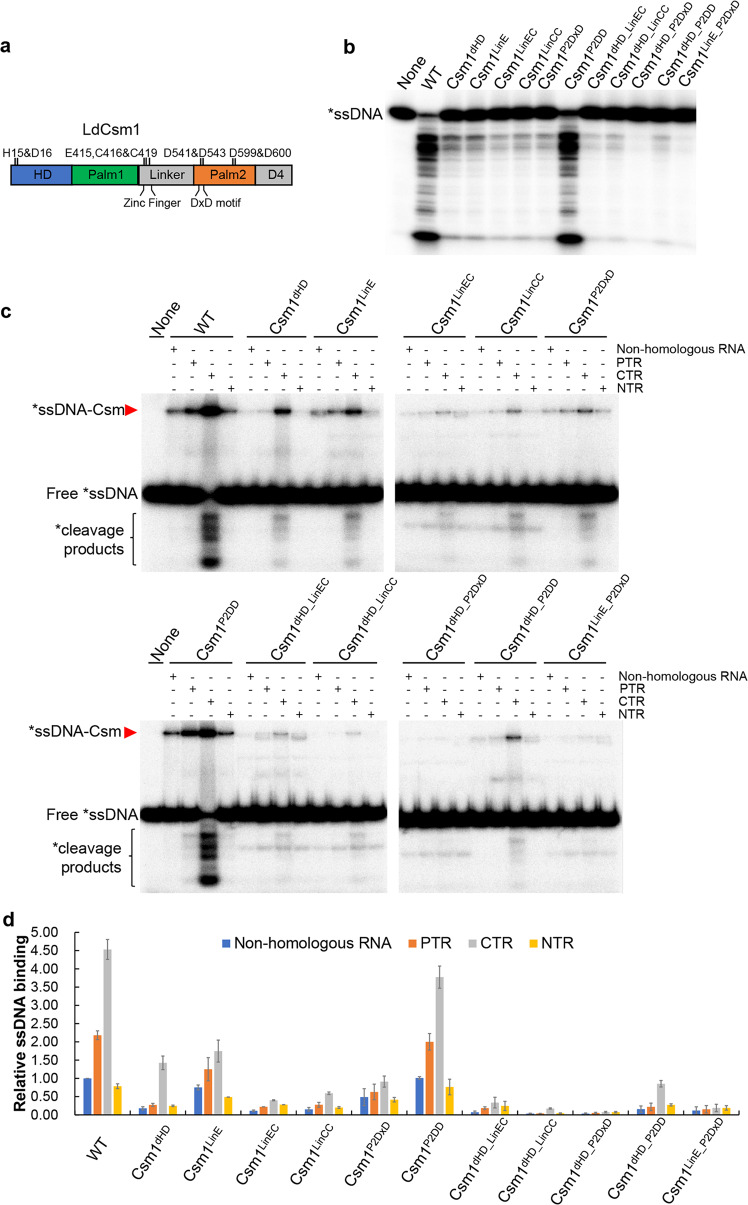
Fig. 4. Effect of LdCsm1 mutations on ssDNA binding and cleavage by the LdCsm effector complex.
a Domain architecture of the LdCsm1 protein. HD represents the HD-type nuclease domain; Palm 1 and Palm 2 denote the two cyclase domains; Linker is a domain that adjoins the Palm1 and Palm2 domains, consisting of four cysteine residues; D4 is located in the C-terminus rich in α-helices. Amino acid residues selected for alanine substitution mutagenesis are indicated with their names and positions. b RNA-activated ssDNA cleavage by effectors carrying one of the constructed LdCsm1 mutants. Fifty nM S10–60 ssDNA substrates were mixed with 50 nM mutated LdCsm carrying each of LdCsm1 mutant proteins and 500 nM CTR and incubated for 10 min. Samples were analyzed by denaturing PAGE. c ssDNA binding by effectors carrying each of the constructed LdCsm1 mutants. Five nM labeled S10–60 ssDNA were incubated for 3 min with 100 nM of LdCsm effectors in the presence of 400 nM of non-homologous RNA (S10 RNA) or 500 nM of one of the target RNAs, PTR or CTR or NTR. Samples were analyzed by non-denaturing PAGE. Red arrowheads indicate the Csm–ssDNA complex. d Relative ssDNA binding between the wild-type LdCsm effector and its LdCsm1 mutated derivatives. The relative ssDNA-binding activities were estimated by image quantification of the non-denaturing PAGE in c by the accessory analysis tool in Typhoon FLA 7000, the ssDNA activity of LdCsm in non-homologous RNA was used as the standard and set up as 1. Results shown are average of three independent assays; bars represent the mean standard deviation (±SD).