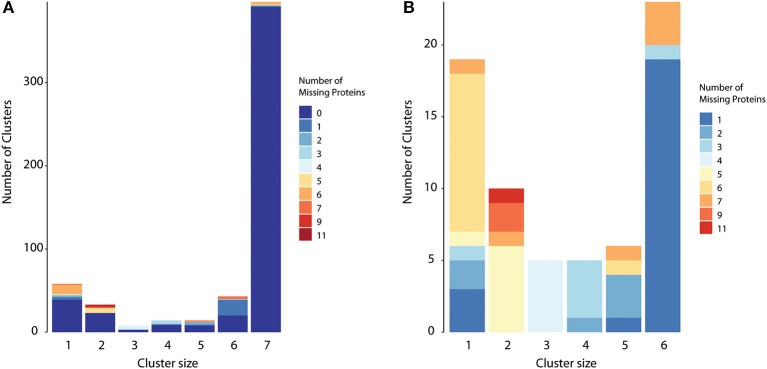
Figure 3.
True core proteins with discrepant surfaceome prediction. To evaluate if clustering could be biased when performed on a smaller dataset, we also executed CMG Biotools in the whole proteome dataset of all strains. Protein composition for most groups was identical regardless if clustering was performed before or after surfaceome prediction (zero missing proteins when clusters are compared) (A). For core proteins (column 7), 98% of the protein groups were identical. For accessory proteins (groups 1–6), this was closer to 55%. When hiding identical clusters (B), it is evident that for most accessory clusters, the number of missing elements adds to the exact number of strains used. This illustrates protein groups with 7 components if no prediction is performed, i.e., true core proteins.