FIGURE 6.
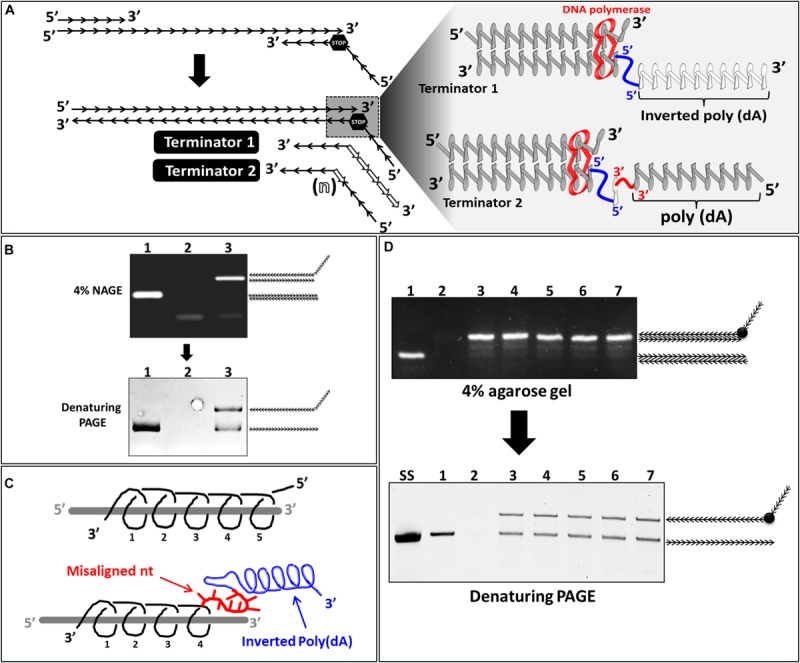
Effect of incorporation of inverted nucleotide/s on ssDNA generation. (A) Schematic illustration of the reverse primer with either an inverted poly(A) tail (terminator 1) or internally inverted nucleotide/s (terminator 2) and the predicted effect of these moieties on DNA backbone and DNA polymerization (magnified). (B) (lane 1): regular PCR product performed on a constant DNA template (100 nt in length); (lanes 2 and 3): products of PCR performed with a reverse primer containing an inverted poly (dA) (Terminator 1) at annealing temperature of 72 and 60°C respectively were analyzed with NAGE (top) and PAGE (bottom). (C) Schematic illustration of the effect of the inverted poly(dA) tail (gray helix) on RP (black helix) backbone structure. (D) NAGE (4% w/v) (left side) and PAGE (8% w/v) (right side) analysis of PCR products performed with reverse primers containing different numbers (1 to 5) internally inverted dAs. (lane 1): product of regular PCR; (lane 2): negative control (no template); (lanes 3 to 7) Products of PCRs performed with RP containing 1, 2, 3, 4, or 5 internally inverted dA/s downstream of a poly(dA) tail (non-inverted) (Terminator 2).
