FIGURE 7.
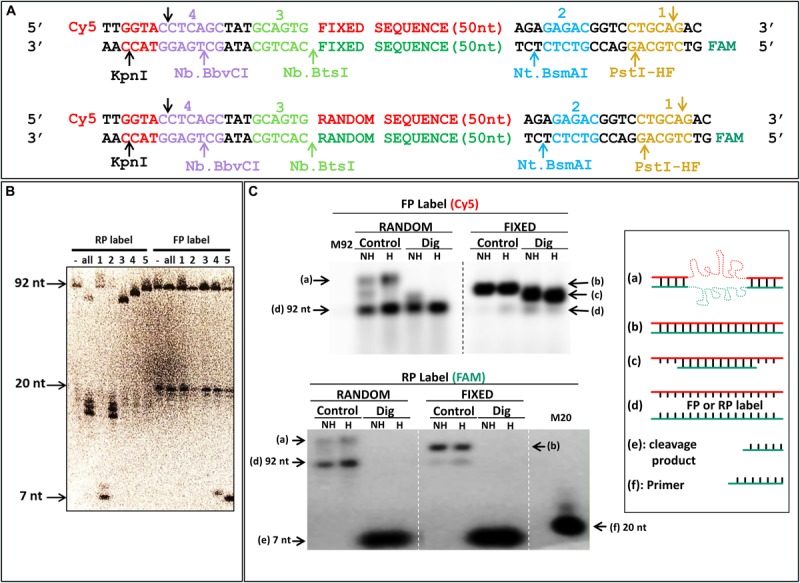
Specific digestion of reverse strand DNA. (A) Scheme of the designed library, and control “fixed” sequence, with cleavage sites of each enzyme pictured in a different color in the constant or random sequence regions with fluorescent labels; “Cy5”: cyanine (Forward); “FAM”: fluorescein (Reverse). (B) Denaturing urea PAGE of digested DNA. Numbered sites in (A) correspond to the enzymes used in the lanes of the gel; RP label and FP label: radioactive labeling of reverse strand via the labeled reverse and forward primers, respectively; “-”: uncleaved PCR; “all”: all three nicking enzymes (2, 3, and 4) together with KpnI or PstI-HF, in RP or FP label, respectively. Full length DNA is 92 bases; the 20 bases band correspond to labeled primers; and 7 bases band to either KpnI (FP) or PstI-HF (RP) cleavage products. (C) Native agarose gel of DNA with fixed and random sequences. Each PCR amplicon was labeled with fluorescence as indicated in (A) and imaged with a Typhoon FLA9500 for Cy5 (FP label) and fluorescein (RP label). M92: marker ssDNA, 92 bases; “M20” Marker ssDNA, 20 bases correspond to labeled primer; “Control”: uncleaved PCR; “Dig”: digestion with all three nicking enzymes (2, 3, and 4) together with PstI-HF, in reverse primer label; full length DNA is 92 bases; and 7 bases band corresponds to PstI-HF (RP) cleavage products; NH: samples were not heated before loading; H: samples were heated to 94°C before loading; “a–f”: different structures of DNA and markers used are schematically represented (on the right side, as in Figure 5). Electrophoresis was done in a room at 37°C in a 3% NAGE (native agarose gel).
