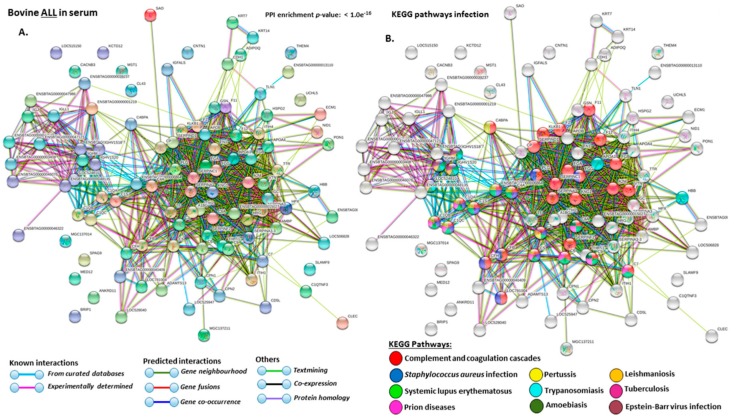
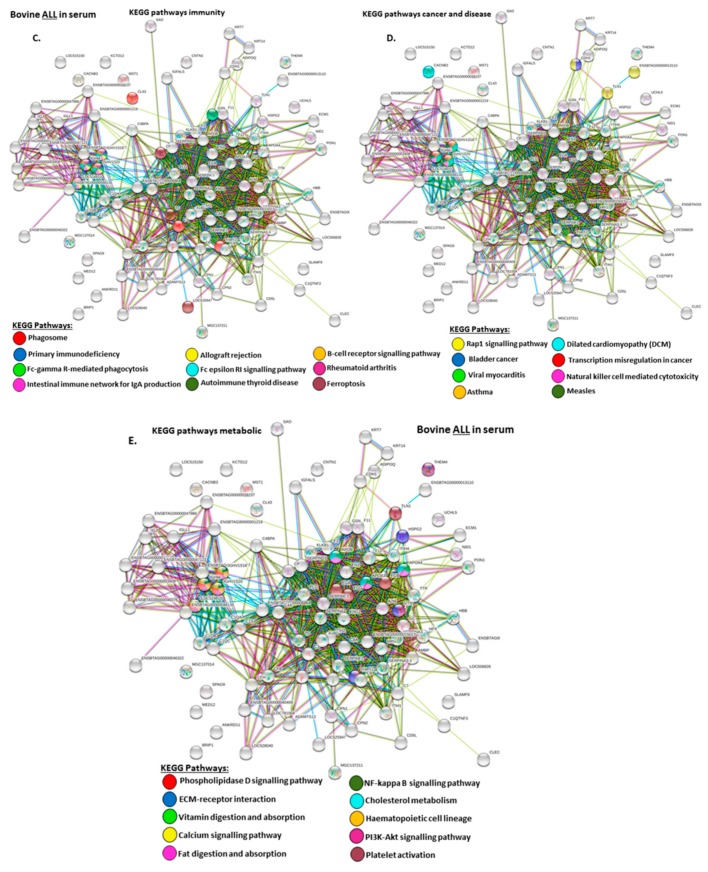
Figure 4.
Protein–protein interaction networks show all deiminated proteins which were identified in bovine serum. Protein–protein interactions were reconstructed based on both known and predicted interactions in serum of Bos taurus, using STRING analysis. (A) Query proteins are indicated by the coloured nodes represent and first shell of interactors. (B) KEGG pathways relating to the identified proteins and reported in STRING and relating to infection are highlighted (see colour code included in the figure). (C). KEGG pathways relating to the identified proteins and reported in STRING are highlighted for immunity (see colour code included in the figure). (D). KEGG pathways relating to cancer and disease for deiminated proteins identified are highlighted (see colour code included in the figure). (E). KEEG pathways relating to metabolism for deiminated proteins identified are highlighted (see the colour code included in the figure). The coloured lines highlight which protein interactions are identified through known interactions (this refers to curated databases, experimentally determined), through predicted interactions (this refers to gene neighborhood, gene fusion, gene co-occurrence) or through co-expression, text mining or protein homology (the colour key for connective lines is included in the figure).