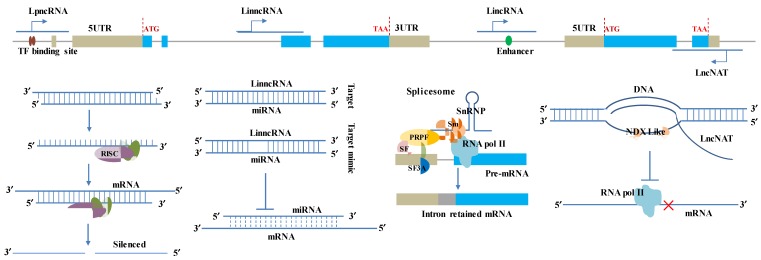
Figure 1.
LncRNA Schematic illustrations for classification and regulation mechanism in functional genes. The lncRNA classes were given on the top based on the DNA sequences and cDNA sequences of two simulated tandem genes. Open reading frames (ORFs) and exons are shown as blue boxes. The introns of the genome sequences are indicated by grey lines. Untranslated regions (UTRs) are shown as beige boxes. The regulation mechanism of lncRNA in functional genes was simulated in the bottom panel, including miRNA precursors, competitor/decoy (miRNA target/target mimic), alternative splicing, and R-loop. The dash lines represent the alignment probability between miRNA and mRNA was reduced due to competition by lncRNAs. The outline and shapes of spliceosome complexes tethering lncRNA are purely schematic; their size and number have no meaning. ‘Sm’ indicates the seven Sm proteins, i.e., B, D1, D2, D3, E, F, and G; ‘PRPF’ and ‘SF’ mean pre-mRNA processing factor and splicing factor; snRNPs contain the indicated lncRNA (containing an Sm site bound to RNA-binding protein) plus stably bound proteins, many of which are shared with the major spliceosome. SnRNP association with RNA polymerase II in the spliceosome has only been inferred to date. The regulated genes could be any potentially functional gene, including pathogen perception and signal transduction-related genes, R genes, their regulator (transcription and splicing factors), and so on.