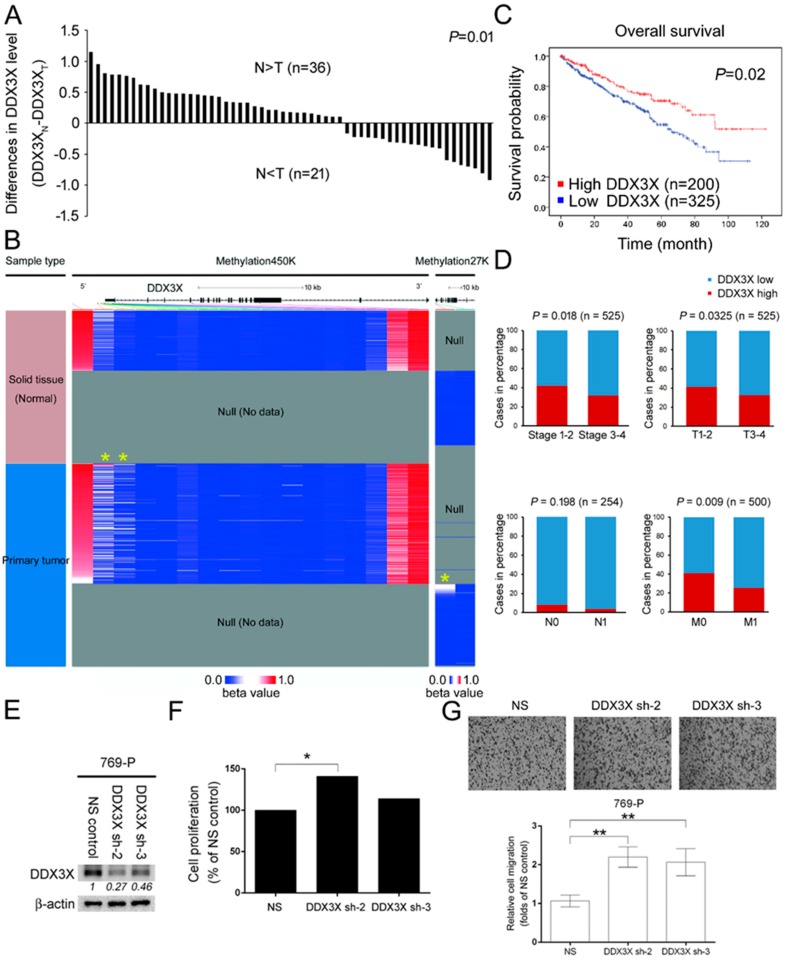
Figure 1.
DDX3X is epigenetically repressed in tumor tissue, and lower DDX3X is correlated with poor overall survival and high tumor-node-metastasis (TNM) status in renal cell carcinoma (RCC) patients. (A) The expression profile of DDX3X in 57 matched renal clear cell carcinomas and adjacent normal tissues was compared. The gene expression profile was measured experimentally using the IlluminaHiSeq_RNASeqV2. Raw data were retrieved from TCGA database and analyzed (Dataset ID: TCGA_KIRC_exp_HiSeqV2_PANCAN). T represents tumor tissue; N represents normal adjacent tissue. The relative difference in DDX3X expression was obtained by DDX3XN-DDX3XT. (B) DNA methylation patterns in solid tissue and primary tumors of the RCC cohort were tested via Methylation450K and Methylation27K platforms and released by The Cancer Genome Atlas (TCGA). The position of the DDX3X protein-coding gene promoter along with the methylation pattern is indicated by a yellow asterisk. The DNA methylation fraction at a specific CpG site was calculated as beta value (β) = M/(M+U+α), where M and U are methylated and unmethylated signal intensities, and α is an arbitrary offset intended to stabilize β values where fluorescent intensities are low. (C) A Kaplan–Meier plot of 525 cancer patients with relatively high and low DDX3X levels. The case numbers in the high and low groups were determined by overall survival. (D) Associations of DDX3X expression with stage, tumor size (T1-4), lymph node (N0-N1) and distant metastasis (M0-M1) in the RCC cohort were analyzed using the χ2 test. (E) The DDX3X expression was silenced by the lentiviral-based transduction of specific shRNA clone 2 and clone 3 in 769-P cells. The protein levels were normalized with corresponding internal controls. The relative DDX3X protein level was shown. NS; non-silencing control. (F) 769-P cell numbers in indicated groups were counted by trypan blue exclusion assay after 24 h of incubation, * p < 0.05. (G) Relative 769-P cell migration upon DDX3X knockdown was evaluated by transwell assay at time point 2.5 h. The p values were represented by **, and considered significant (p < 0.01).