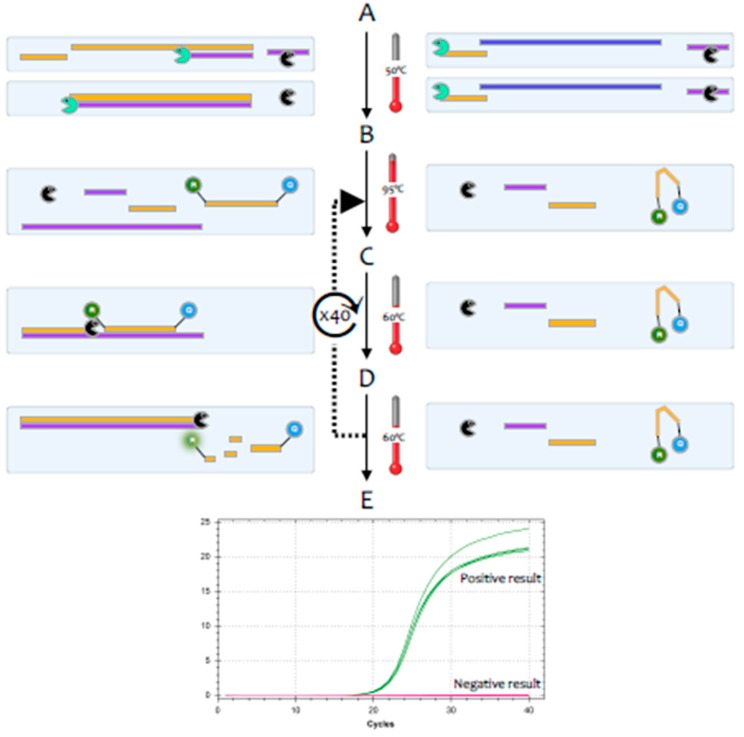
Figure 2.
Signal generation during a RT-qPCR test. Test reagents include a buffer, both enzymes, target-specific DNA primers, and a target-specific DNA probe that is labelled at one end with a fluorescent label and at the other with a quencher. Samples on the left and right contain the same primers and probe, but the one on the left harbours target RNA, whereas the one on the right does not. A. RT: Samples are incubated at around 50 °C, which results in the RT transcribing target-specific cDNA from one of the strand-specific primers on the left, with no reverse transcription on the right. B. Denaturation: Samples are heated to 95 °C, which denatures the RNA but leaves the cDNA intact. C. Annealing: the temperature is lowered to around 60 °C, with the actual temperature assay-dependent. This allows both the target-specific primers and probe to bind to their respective targets on the left, whereas primers and probe remain unbound on the right. D. Polymerisation: this step may be combined with the annealing step. On the left, the polymerase extends DNA synthesis, initially from one primer only, but after the first cycle from both, and displaces and hydrolyses any bound probe. This separates fluorophore and quencher and results in the emission of light if the fluorophore is excited at the appropriate wavelength. On the right, none of this occurs, and no light is emitted. This first cycle is followed by a further, user-defined number of cycles, indicated by the stippled arrow leading back to step B. E. Amplification plots obtained for each sample track the increasing emission of light characteristic of a positive result from the sample on the left (green plot), whereas the sample with no amplifiable target on the right records no light emission and a negative result (red plot).