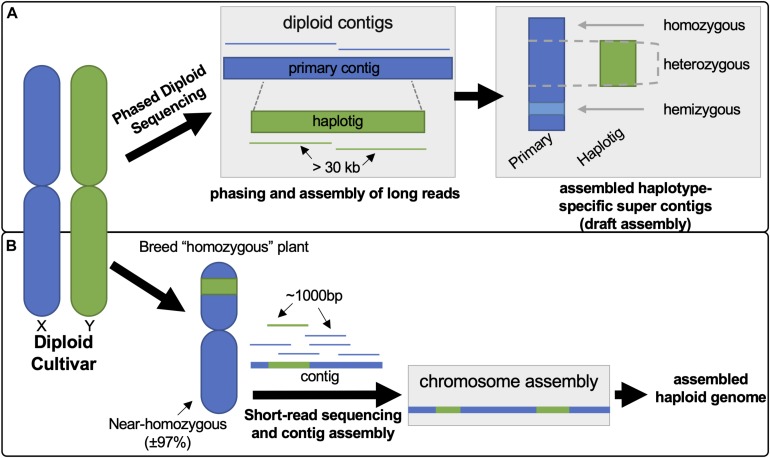
FIGURE 1.
Illustration contrasting the differences in approach between phased diploid (A) and short-read sequencing (B) to generate the diploid and PN40024 reference genomes, respectively. (A) PacBio long-reads (> 30 kb) undergo phasing during the assembly, using FALCON-UNZIP, resulting in the generation of a pseudomolecule known as a primary contig. This primary contig region represents both possible haplotypes. The phasing algorithm identifies reads with high heterozygosity to call an alternate genomic region (haplotig) which is assembled separately allowing for the generation of haplotype regions. (B) The reference genome was generated from an inbred clone of Pinot Noir (PN40024), reducing genomic complexity to near homozygosity. Assembly of the short-reads, generation of contigs and subsequent mapping/assembly to chromosomes lead to the PN40024 reference genome.