Extended Data Fig. 4. Cluster annotation, supporting evidence for HSC self-renewal and Lineage-negative, c-Kit+ cells validation in Tet2 KO.
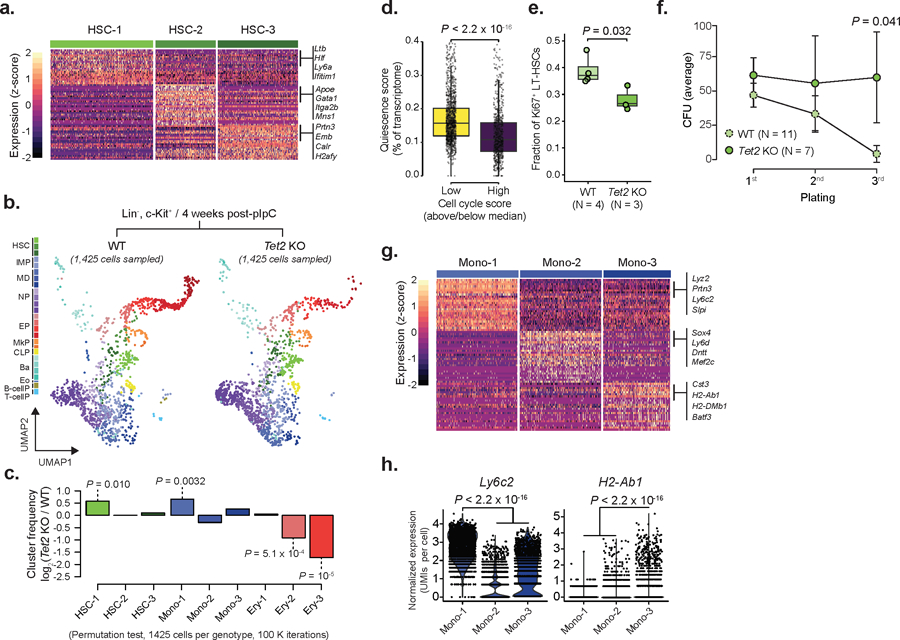
a) Differentially expressed genes for WT cluster HSC-1 (492 cells), HSC-2 (288 cells) or HSC-3 (384 cells), relative to the remaining HSC clusters from Chromium data (n = 4 mice; logistic regression with Bonferroni correction; FDR < 0.05). b) Drop-seq data for Lin−, c-Kit positive cells for WT (2,986 cells, n = 2 mice) or Tet2 KO (1,425 cells, n = 2 mice) progenitors 4 weeks after recombination (HSCs = Hematopoietic stem cells; IMP = Immature myeloid progenitors; MD = Monocytic-dendritic progenitors; NP = Neutrophil progenitors; EP = Erythroid progenitors; MkP = Megakaryocyte progenitors; CLP = Common lymphoid progenitors; Ba = Basophil progenitors; Eo = Eosinophil progenitors; B-cellP = B-cell progenitors; T-cellP = T-cell progenitors). c) Frequency changes for HSCs, MDs and EPs 4 weeks after recombination (Permutation test on 1,425 randomly sampled cells from each genotype, with 105 iterations). d) Quiescence score per cell cycle category (above/below median) in WT HSCs (n = 1,982 cells; two-sided Wilcoxon rank sum test). e) Flow cytometry of cell cycle in LT-HSCs as measured by Mki67 expression for WT (n = 4 mice) or Tet2 KO (n = 3 mice) 4 weeks after recombination (two-sided Student t-test). f) Serial re-plating colony-formation assays for WT (n = 11) and Tet2 KO (n = 7) Lin−, c-Kit+ bone marrow hematopoietic (CFU = colony formation unit; dots represent the mean; error bars represent standard deviation; two-sided Students t-test). g) Differentially expressed genes per WT cluster Mono-1 (n = 344 cells), Mono-2 (n = 345 cells) or Mono-3 (n = 284 cells), relative to the remaining monocyte clusters. Differentially expressed genes were defined from Chromium data (n = 4 mice; logistic regression with Bonferroni correction; FDR < 0.05). h) Expression of Ly6c2 and H2-Ab1 in WT Mono-1 (n = 344 cells), Mono-2 (n = 345 cells) and Mono-3 (n = 284 cells) clusters (logistic regression with Bonferroni correction).
