Extended Data Fig. 5. Flow cytometry validation, peripheral blood counts, in vitro colony-forming assay and 20 weeks post pIpC validations of Tet2 KO frequency changes.
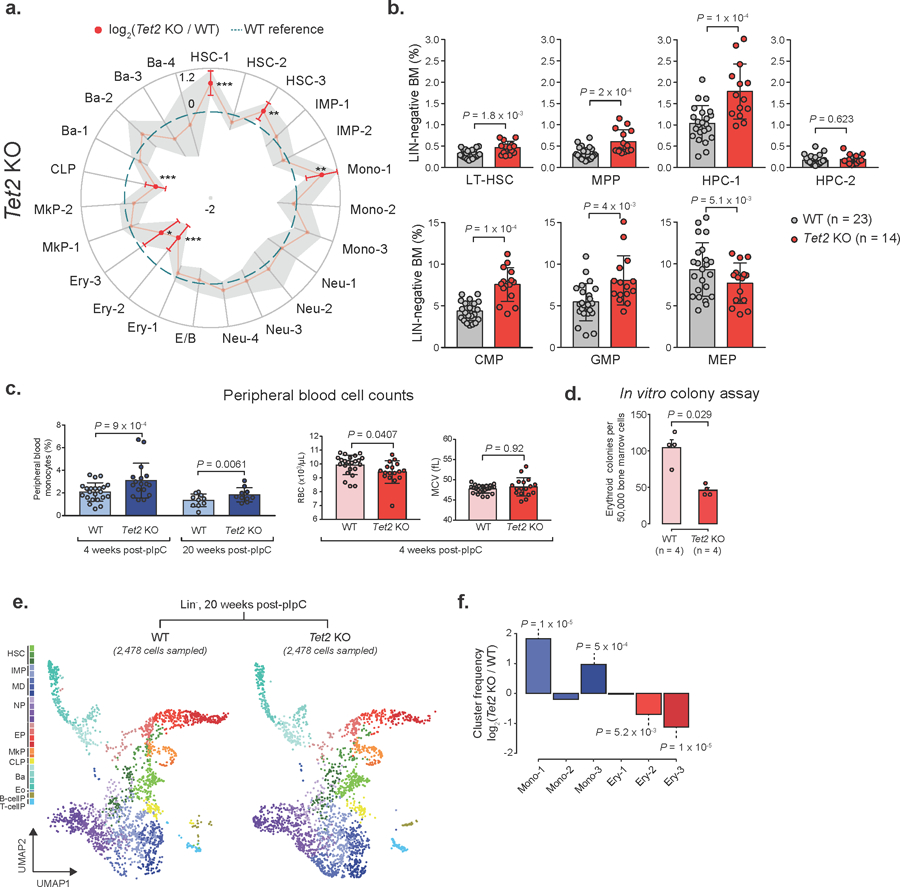
a) Frequency changes for Lin− Tet2 KO (18,651 cells, n = 7 mice) relative to WT (17,702 cells, n = 7 mice) 4 weeks after recombination. Red dots indicate significant frequency changes; red error bars represent standard deviation; dashed line indicates WT reference frequencies; grey shadow region indicates +/− standard deviation (LMM followed by ANOVA; * P < 0.05; ** P < 0.01; *** P < 0.001). b) Flow cytometry for WT (n = 15) and Tet2 KO (n = 23) mice 4 weeks after recombination (two-sided Students t-test; bars represent the mean value, error bars represent the standard deviation; LT-HSC = long-term hematopoietic stem cell; MPP = Multi-potent progenitor; HPC = Hematopoietic progenitor cell; CMP = common myeloid progenitor; GMP = Granulocyte-monocyte progenitor; MEP = megakaryocyte-erythrocyte progenitor). c) Peripheral blood cell counts from WT (n = 10) or Tet2 KO (n = 10) mice, either 4 or 20 weeks after Cre-mediated recombination (two-sided Students t-test; bars represent the mean and error bars represent the standard deviation. Each dot represents a mouse replicate; RBC = red blood cells; MCV = mean corpuscular volume). d) Erythroid colony-forming assay for WT (n = 4) or Tet2 KO (n = 4) mice, 4 weeks after recombination (two-sided Student t-test; bars represent the mean number of colonies for each genotype; error bars represent standard deviation). e) Drop-seq data showing 2,478 randomly sampled cells from Lin− cells for WT (2,757 cells) or Tet2 KO (2,875 cells) 20 weeks after recombination (HSCs = Hematopoietic stem cells; IMP = Immature myeloid progenitors; MD = Monocytic-dendritic progenitors; NP = Neutrophil progenitors; EP = Erythroid progenitors; MkP = Megakaryocyte progenitors; CLP = Common lymphoid progenitors; Ba = Basophil progenitors; Eo = Eosinophil progenitors; B-cellP = B-cell progenitors; T-cellP = T-cell progenitors). f) Frequency changes for monocyte (Mono 1–3) and erythroid (Ery 1–3) progenitor clusters (permutation test).
