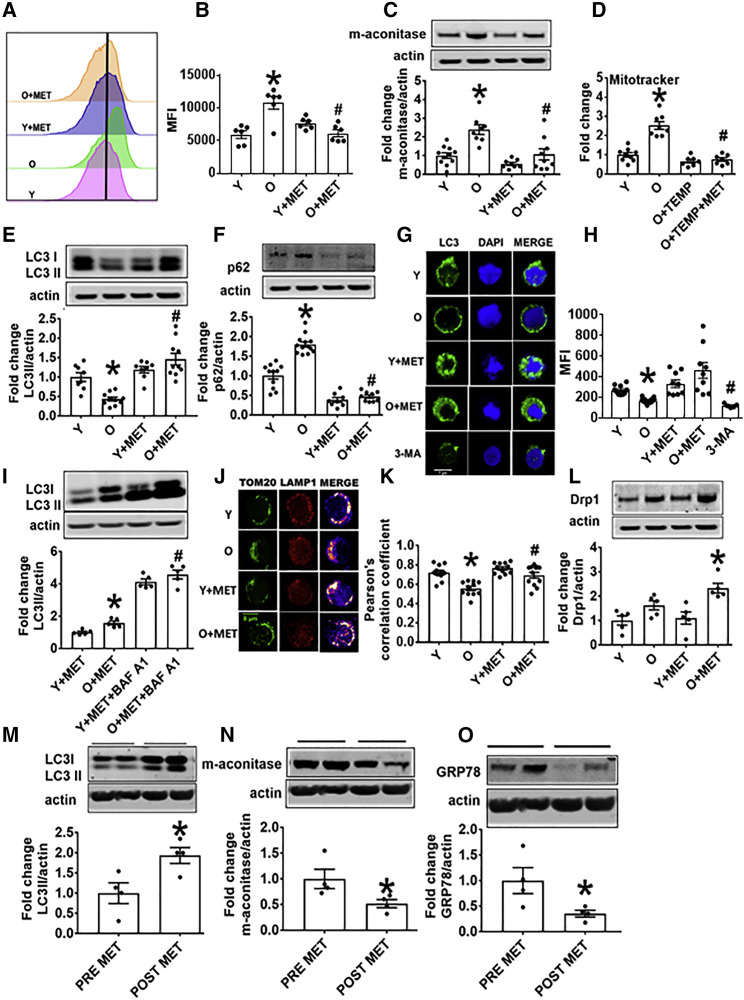
Figure 4.
Metformin Promotes Mitochondrial Turnover and Mitophagy in CD4+ T Cells from O Subjects
(A and B) MitoTracker green fluorescence in CD4+ T cells from O and Y subjects assessed by flow cytometry. n = 6.
(C) Mitochondrial matrix protein m-aconitase in CD4+ T cells from O and Y subjects as measured on western blots. n = 7–10.
(D) Mitochondrial mass assessed via Mitotracker green fluorescence in the presence of Tempol (TEMP) ± MET as indicated. n = 7–8.
(E and F) Expression of the autophagy proteins LC3II (E) or p62 (F) in cells, measured on western blots. n = 8–10.
(G and H) Autophagosome formation as indicated by puncta and quantitated by confocal microscopy. 3-MA is an inhibitor or autophagy thus serves as a negative control. n = 3.
(I) LC3II expression in CD4+ T cells as indicated, following treatment with metformin + BAF A1 as a positive control for autophagy. n = 5.
(J and K) Localization in representative CD4+ cells (J), and quantitation of co-localization of the mitochondrial protein TOM20 and the lysosomal protein (LAMP1) ± metformin as indicated (K). n = 4 with multiple dots from some N’s shown.
For both confocal analyses (G, H, J, and K), 3 cells/field and 3 fields/slide were imaged using 63× oil immersion in Zeiss microscope. The average fluorescence/field is reported.
(L) Expression of mitochondrial fission protein Drp 1 on western blots. n = 5.
(M–O) Indicators of autophagy (M) LC3II, (N) m-aconitase, or (O) GRP78 quantified in CD4+ T cells from pre-diabetes subjects sampled before or after 3 months’ administration of metformin (1,000 mg/day; n = 4). ∗p < 0.05 versus Y or pre-met, #p < 0.05 versus O.
Data are represented as mean ± SEM. Fold change is compared to either Y, Y + MET, or PRE-MET.