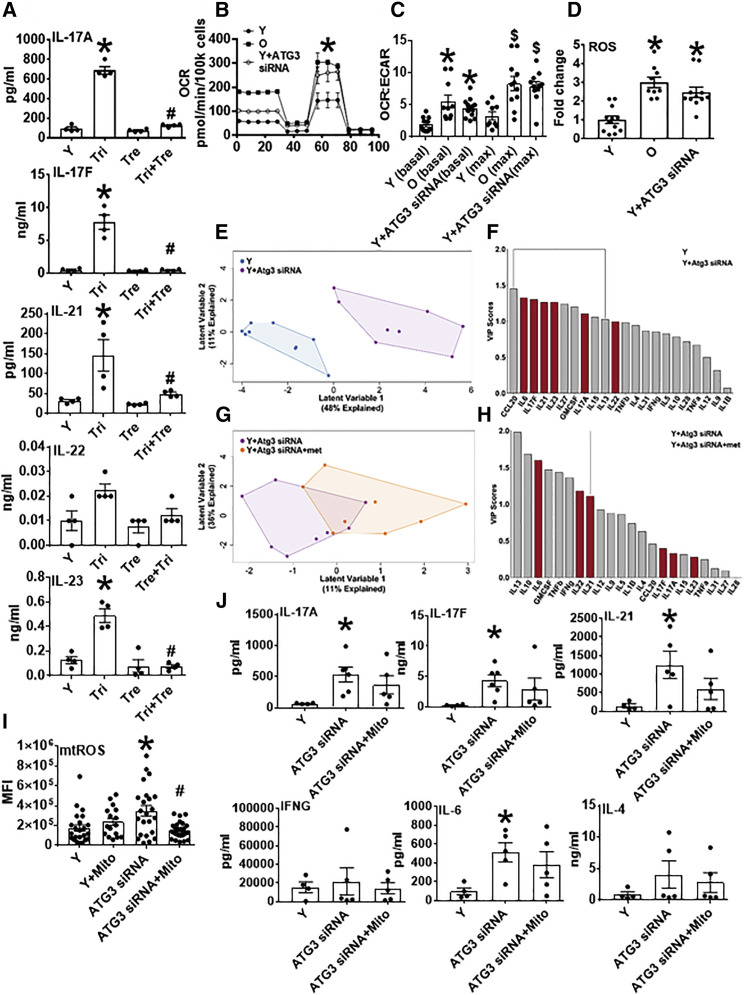
Figure 5.
Genetic Inhibition of Autophagy Recapitulates Respiratory Profiles of Cells from O Subjects
(A) Th17-associated cytokine production by CD4+ T cells from Y subjects, with cells stimulated 40 h αCD3/αCD28 in the presence of trimetazidine, a fatty acid oxidation inhibitor; alone; or in combination with trehalose, an autophagy activator. n = 4. ∗p < 0.05 versus Y by one-way ANOVA.
(B) Mito stress test XF profiles from 40 h αCD3/αCD28-stimulated CD4+ T cells from Y or O subjects as indicated. Autophagy dysfunction was induced in cells from Y subjects using siRNA-mediated ATG3 knockdown. n = 8–12.
(C and D) OCR:ECAR ratio (C) and ROS generation (D) measured by DCFDA in CD4+ T cells manipulated as indicated. n = 8–12.
∗p < 0.05 versus Y by SHORE (Nicholas et al., 2017) (B) or one-way ANOVA (C and D). ∗p < 0.05 versus Y (basal), $p < 0.05 versus Y (max) (C).
(E and G) PLSDA analysis differentiated combinatorial “inflammation” of CD4+ cells from Y subjects (blue), Y with siRNA-induced autophagy dysfunction (purple), or autophagy dysfunction and metformin (met; orange). n = 8–9.
(F and H) VIP scores rank cytokines important for differentiating data clouds in (E) and (G). Comparison of Figure 5F with 1D highlights profiles that differentiate Y from either O or Y + ATG3 siRNA conditions. n = 8–9.
(I) Mitochondrial ROS generation measured by MitoSOX in CD4+ T cells manipulated as indicated; 3 cells/field and 4 fields/slide were imaged using 40× in Zeiss microscope. The average fluorescence/field is reported. n = 4–5.
(J) Cytokine production by CD4+ T cells from young subjects, with cells stimulated 40 h αCD3/αCD28 after autophagy inhibition and in the presence of mitoTEMPO, a mitochondrial ROS-specific scavenger (1 μM, added 3 h post αCD3/αCD28 stimulation). n = 5–6.
(A, C, D, I, and J) Data show mean ± SEM. Fold change is compared with Y.
See also Figures S4 and S5.