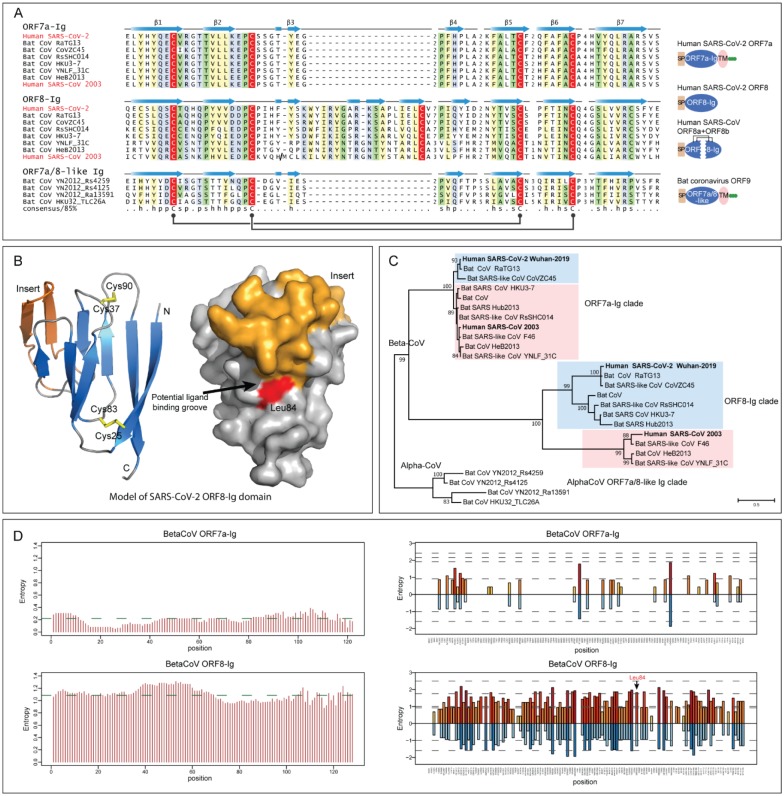
Figure 2. Sequence, structure and evolutionary analysis of novel Ig domain proteins in SARS-related CoVs.
(A) Multiple sequence alignment (MSA) and representative domain architectures of ORF7a-Ig, ORF8-Ig, and ORF7a/8-like Ig domain families. Each sequence in the MSA was labelled by its species abbreviation followed by its source. The predicted secondary structure is shown above each alignment and the consensus is shown below the super-alignment, where h stands for hydrophobic residues, s for small residues, and p for polar residues. Two pairs of conserved cysteines that form disulfide bonds are highlighted in red. (B) Homology model of SARS-CoV-2 ORF8-Ig domain (YP_009724396.1) and the location of the hypervariable position corresponding to Leu84 in the predicted ligand-binding groove. The β-sheets of the common core of the Ig fold are colored in blue, the insert in ORF8-Ig in orange and the loops in grey. The characteristic disulfide bonds are highlighted in yellow. (C) Maximum likelihood phylogenetic analysis of CoV Ig domain families. Support values out of 100 bootstraps are shown for the major branches only. (D) Entropy plot for the ORF7a and ORF8 proteins in betacoronavirus. Left: Shannon entropy computed for each column for a character space of 20 amino acids and presented as mean entropy in a sliding window of 30 residues. The mean entropy across the entire length of the protein is indicated as a green horizontal line. Right: Shannon entropy in regular amino acid alphabet (20 amino acids) are shown above the zero line in shades of orange. Shannon entropy in a reduced alphabet of 8 residues are shown below the zero line in shades of blue. If a position shows high entropy in both alphabets it is a sign of potential positive selection at those positions for amino acids of different chemical character.