Figure 1. MS Crosslink Analysis of Sin3/HDAC Complexes.
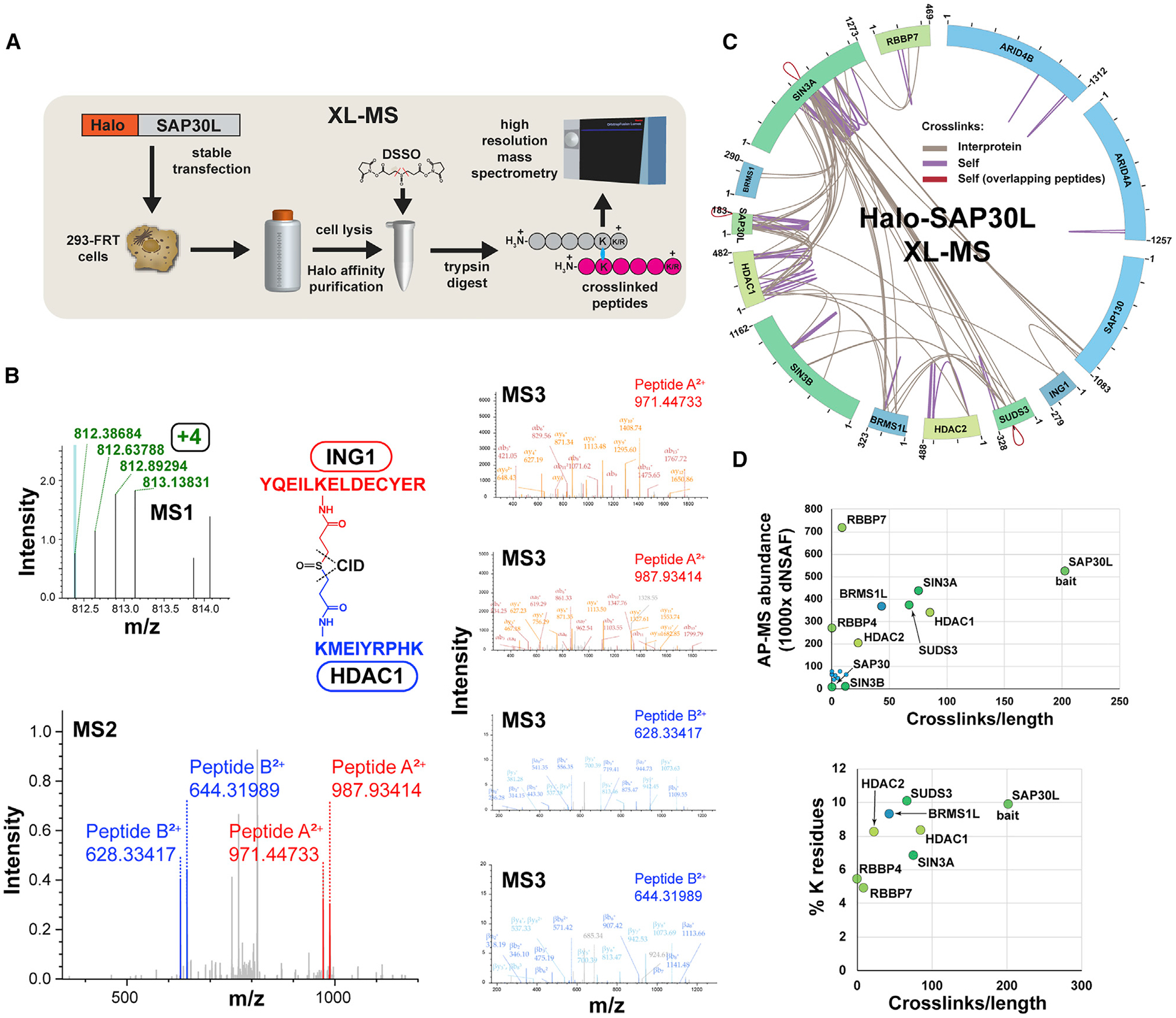
(A) Workflow for XL-MS experiments. In contrast to AP-MS experiments, Halo-purified samples were treated with DSSO before analysis by high-resolution mass spectrometry.
(B) High-resolution MS2 and MS3 spectra used to identify the ING1-HDAC1 crosslinked peptide. Putative crosslinked peptides with charge ≥ +4 were selected during MS1 analysis and low-energy collision-induced dissociation (CID) was used to cleave the DSSO crosslinker, generating a pair of fragments for each peptide (MS2-ING1 peptides shown in red; HDAC1 in blue). The four fragments were sequenced using MS3.
(C) Crosslink map for the Sin3/HDAC complex. Crosslink identifications are from three XL-MS experiments. Values indicate protein length (amino acids). Details of crosslinks are in Table S2.
(D) Relationship between observed crosslink abundance and either protein abundance or lysine content. Crosslink abundance is 1,000 × (semi-crosslinks/protein length [aa]), with two semi-crosslinks counted for each of the protein’s self-crosslinks and one for each of the protein’s interprotein crosslinks. Protein abundance distributed normalized spectral abundance factor (dNSAF) values for SIN3 subunits co-purifying with Halo-SAP30L (four biological replicates) were published previously (refer to Table S3 in Banks et al., 2018).
