Abstract
Background and Purpose:
Culture-based identification methods have been the gold standard for the diagnosis of candidal onychomycosis. Molecular technologies, such as polymerase chain reaction (PCR) assays, can provide an alternative for the rapid detection of Candida species. The present study was conducted to investigate a pan-Candida PCR assay based on the translation elongation factor 1-alpha (TEF-1α) gene for the detection of the most prevalent pathogenic Candida species.
Materials and Methods:
For the purpose of the study, an optimized pan-Candida PCR primer pair was designed, and the target was amplified and sequenced. The analytical and clinical diagnostic performance of the designed primers was tested using 17 reference strains, 137 nail scrapings suspected of onychomycosis, and 10 healthy nail specimens.
Results:
The use of the universal pan-Candida primers designed on TEF-1α gene resulted in the successful amplification of a 270-base pair fragment in all Candida species tested, except for C. glabrata, and reacted neither with other fungi nor with E. coli. The sequence difference count matrix showed poor insertion/deletion differences (0-2 nt) among Candida species. Among 137 nail specimens, 35% (n=48), 30.7% (n=42), and 40.1% (n=55) of the samples were found to be positive by direct microscopy, culture, and pan-Candida PCR, respectively.
Conclusion:
Based on the findings, the PCR-based detection targeting the DNA TEF-1α gene is a rapid and simple procedure for the diagnosis of candidal onychomycosis directly from nail sample.
Key Words: Candidal onychomycosis, Pan-Candida PCR assay, Translation elongation factor 1-alpha
Introduction
Candida species are common human commensal and important human yeast pathogens that cause a wide spectrum of diseases ranging from less severe superficial lesions to life-threatening systemic conditions in a vast spectrum of immunocompromised patients [1]. The incidence of infections due to Candida species has increased markedly as a result of a growing number of immunocompromised and critically ill patients [2]. Candida contains about 150 species. However, only a few of these species, including C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. dubliniensis, and C. krusei, are implicated in human pathogenesis.
Superficial candidiasis may involve one or more mucocutaneous tissues, including the skin, mucous membrane, and nails [3]. Nail infections caused by Candida species known as candidal onychomycosis are normally associated with paronychia, onychia, or chronic mucocutaneous candidiasis. Further comprehensive studies have pointed out that Candida is the second most common etiology for onychomycosis after dermatophytes (tinea unguium) [4]. Conventional laboratory techniques for the diagnosis of candidal onychomycosis, such as direct microscopic investigation and mycological culture, are time consuming and laborious. Recent studies have shown that advances in molecular technologies are useful in detecting microbial infections without the need for microbial isolation [5, 6].
Nuclear ribosomal DNA (rDNA) regions, including ITS1 and ITS2, have been the most commonly used targets for the detection/identification of Candida species [7, 8]. Nevertheless, the description and characterization of new genetic markers for Candida species can clarify its taxonomy and might be helpful for detection/identification goals. The protein-coding genes (e.g., translation elongation factor 1-alpha [TEF-1α]) have been proven to be a powerful tool for the species delimitation of the taxonomically complex Fusarium species, considered an alternative to rDNA for species identification [9, 10]. Likewise, the gene is useful for developing robust phylogenetic inferences for other groups of pathogenic fungi, such as zygomycetes [11], dermatophytes [12], and Aspergillus species [13]. Since TEF-1α gene has not been used in Candida taxonomy and detection, our aim was to investigate TEF-1α as a new genetic marker to evaluate a pan-Candida PCR assay in the diagnosis of candidal onychomycosis.
Materials and Methods
Standard strains and clinical specimens
The specificity and sensitivity of the designed PCR system were determined against 17 standard strains of fungi. These strains included C. albicans (ATCC 5982, ATCC 2730, ATCC 562, ATCC 1912 and ATCC 64553), C. tropicalis (CBS 94 and ATCC 750), C. krusei (ATCC 6258), C. glabrata (ATCC 90030 and ATCC 863), C. dubliniensis (ATCC 8500 and ATCC 7987), C. parapsilosis (ATCC 4344 and ATCC 90018), Cryptococcus neoformans (ATCC 9011), A. flavus (ATCC 64025), A. fumigatus (ATCC 14110), and environmental isolates of penicillium chrysogenum and Fusarium solani. An standard isolate of Escherichia coli (ATCC 43894) was evaluated as a negative control.
A total of 137 nail scrapings collected from patients who referred to the Medical Mycology Laboratory of Razi Hospital in Tehran [14] were used as clinical samples. Nail specimens from 10 healthy volunteers were used as negative clinical controls to verify the diagnostic specificity of the tests. All specimens were examined by both direct microscopy and culture, and a portion of each sample was stored at -20°C for use in the PCR assay. This research was approved by the Ethics Committee of the Shiraz University of Medical Science (IR.SUMS.REC.1398.020).
Mycological examination
A part of the samples was observed with 20% KOH, and the other part was inoculated onto three different points on Sabouraud dextrose agar (SDA) plate (Merck, Darmstadt, Germany) containing chloramphenicol (0.05 mg/l), with and without actidione (500 mg/l) (Merck, Darmstadt, Germany). The inoculated specimens were incubated at 28°C for 1-4 weeks. The culture plates were examined for fungal growth twice a week.
Primer design
The sequences of TEF-1α in Candida species and other causative agents of onychomycosis, such as dermatophytes and non-dermatophytic molds, were downloaded from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih. gov/pubmed/). The number of sequences for Candida species is limited in NCBI (a total of 13 TEF-1α sequences representing 5 species of Candida). Region of sequences that were conserved within Candida species but different from other fungi responsible for onychomycosis was chosen manually to design a pan-Candida primer pair (Table 1) considering the criteria specified by the Primer Biosoft [15] and optimized using the Multiple Primer Analyzer) https://www.thermofisher.com). The primers were also synthesized by the Bioneer Company (Bioneer, Korea).
Table 1.
Standard and clinical strains of fungi sequences used in this survey for sequence analysis of translation elongation factor 1-alpha gene in silico studies
| Species | Strain | EF-1α accession no. | Size of EF-1α partial sequence (bp) |
|---|---|---|---|
| Candida albicans | ATCC18804 | DQ4472251 | 742 |
| ATCC 18804 | KC5074361 | 929 | |
| SC5314 | XM7050562 | 1377 | |
| SC5314 | M299351 | 2369 | |
| SC5314 | M299341 | 2411 | |
| Candida tropicalis | ATCC 13803 | DQ4472421 | 742 |
| MYA-3404 | XM0025474801 | 1377 | |
| MYA-3404 | XM0025488371 | 1377 | |
| Candida parapsilosis | ATCC90018 | DQ4472381 | 742 |
| Candida dubliniensis | NCPF 3949 | DQ4472291 | 742 |
| CD36-07890 | XM002417390 | 1377 | |
| CD36-22520 | XM002418681 | 1377 | |
| Candida glabrata | ATCC 66032 | DQ4472291 | 742 |
| Epidermophyton floccosum | Clinical sample | KM6781911 | 732 |
| Clinical sample | KM6781881 | 732 | |
| NBRC 9045 | KM6780601 | 732 | |
| Microsporum canis | NRBC 9182 | JN6629361 | 720 |
| CBS 130811 | KM6780521 | 719 | |
| mums138 | MG3569341 | 648 | |
| Microsporum gypseum | CBS 174.64 | KM6780691 | 733 |
| IFO 8228 | KM6780571 | 748 | |
| CBS 130820 | KM6781611 | 748 | |
| Trichophyton interdigital | JCM 1891 | KM6780581 | 766 |
| CBS 130788 | KM6781421 | 768 | |
| CBS 130940 | KM6781731 | 767 | |
| Trichophyton rubrum | CBS 130927 | KM6780551 | 737 |
| DSM 107606 | MH8025051 | 644 | |
| DSM 107668 | MH8025061 | 624 | |
| Trichophyton tonsurans | CBS 130822 | KM6780541 | 756 |
| mums73 | MG3568841 | 667 | |
| mums57 | MG3568801 | 669 | |
| Aspergillus fumigatus | AX 2102 I | KJ4764101 | 873 |
| JCM 10253 | KM9219611 | 674 | |
| CBS 101640 | KM9219681 | 612 | |
| Aspergillus flavus | CBS 573.65 | KM9219761 | 663 |
| JCM 2061 | KM9219751 | 684 | |
| JCM2061 | KM921975.1 | 684 | |
| Aspergillus niger | ITEM 7090 | FN6654031 | 758 |
| ITEM 7496 | FN6656571 | 758 | |
| DUCC5023 | KJ6382501 | 579 | |
| Fusarium solani | VI04903 | FN6898261 | 691 |
| dq240 | KR0255571 | 750 | |
| Fusarium oxysporum | NRRL 45915 | FJ9854261 | 630 |
| NRRL 38544 | FJ9854081 | 629 | |
| NRRL 46589 | FJ9854381 | 631 | |
| Acremonium recifei | CBS 188.82 | KP0126471 | 763 |
| CBS 541.89 | KP0126501 | 683 | |
| CBS 220.84 | KP0126481 | 554 | |
| Scopulariopsis brevicaulis | UTHSC 06-619 | HG3803651 | 957 |
| UTHSC 06-1072 | LM6525861 | 957 | |
| UTHSC 09-1373 | LM6525871 | 967 | |
| Penicillium chrysogenum | AC4503 II | KJ4764051 | 810 |
| AL0202 | KJ4764061 | 921 | |
| MOS731 | KP0090001 | 990 |
Preparation of genomic DNA
DNA was extracted and purified from the fungal colonies using the boiling method [16]. Briefly, a small amount of fresh colony was suspended in 200 µL of distilled water, boiled at 95C for 15 min, and centrifuged for 4 min at 7,000 rpm; subsequently, the supernatant was used for PCR. For clinical specimens, DNA was extracted by means of a commercial kit (Yekta Tajhiz Azma, Iran) according to the manufacturer’s protocol after being treated by 500 μL of a digestion buffer (containing 400 mM Tris-HCL [pH=8], 60 mM EDTA [pH=8], 150 mM NaCl, and 1% sodium dodecyl sulfate) for 30 min and crushed for 3 min by a mechanical grinder.
Polymerase chain reaction
The PCR reactions were performed using 2X PCR Master Mix (Amplicon, Denmark), 0.5 μM of each primer, 4 μL of DNA template, and sufficient distilled water to reach a final volume of 25 μL. The amplification was accomplished using one cycle at 95°C for 3 min, as well as 35 cycles at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, followed by a final extension step at 72°C for 3 min. Negative (i.e., sterile distilled water) and positive (i.e., C. parapsilosis [ATCC 4344]) controls were included in each extraction run. Furthermore, 5 μL of the aliquots of the amplicons was electrophoresed using a 1.1% agarose gel in Tris-Borate-EDTA buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA, and pH of 8.3) and visualized under ultraviolet irradiation after ethidium bromide staining. A 100-bp DNA ladder was used as a molecular size marker.
DNA Sequence Analysis
The PCR product of standard Candida species examined in this study was purified and sequenced unilaterally with the forward primer on an automated DNA sequencer (ABI PrismTM 3500 Genetic Analyzer, Genetic Group) and edited with the Geneious (http://www.geneious.com) software. The sequences were compared with those in the Genebank database using the web-based software BLAST, (http://www.ncbi.nlm.nih.gov/BLAST). The sequence of each sample was subjected to ClustalW pairwise and multiple alignments using the MEGA6 software, and a phylogenetic tree was built using the maximum-likelihood algorithm with the Tamura-Nei parameter.
Analytical sensitivity of primers
To determine the detection limit of this system, fungal inocula were prepared from culture for C. albicans (ATCC 5982). The inocula were adjusted to a turbidity of 0.5 on the McFarland scale, with approximately 1-5×106 CFU/ml, counted in a hemocytometer. The inoculum was diluted with sterile saline at a concentration of 101-106 CFU/ml. Subsequently, 200 μL of pool nail samples made from healthy nails added to each dilution. Commercial kit (Yekta Tajhiz Azma, Iran) was applied for DNA extraction from the nails treated with fungi. Eventually, PCR was performed on DNAs extracted from six serial dilutions and positive and negative controls.
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the pan-Candida PCR against the culture (as the reference method). The method of chi square test was used for categorical data. Statistically, the P-value<.05 was considered as significant. The data were analyzed using SPSS software Version 22.0.
Results
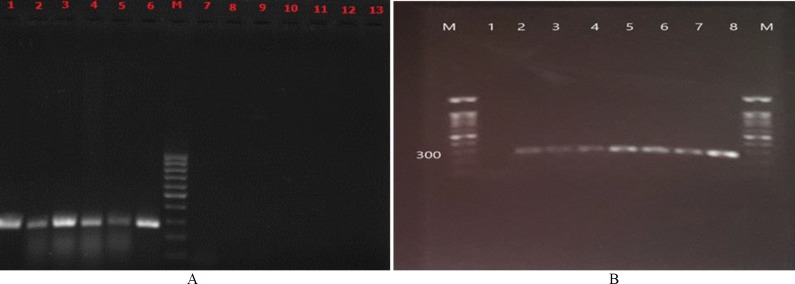
By aligning the TEF-1α gene sequences of most of the common Candida species, a highly conserved region was used for designing a new pan-Candida pair primer for PCR detection. The primers included CEFF: 5'-GGACAAAAACAGATTTGAA-3' as forward primer and CEFR: 5'-TTGCAATGGCAATCTCAAT-3' as reverse. The primers amplified an approximately 270-bp Candida-specific fragment in all tested Candida species, except for C. glabrata and reacted neither with other fungi nor with E. coli DNAs (Figure 1 left).
Figure 1.
(A) Electrophoresis patterns of pan-Candida polymerase chain reaction using DNA template; lane 1) Candida albicans (ATCC 5982), lane 2) Candida tropicalis (ATCC 750), lane 3) Candida parapsilosis (ATCC 4344), lane 4) Candida albicans (ATCC 64553), lane 5) Candida tropicalis (CBS 94), lane 6) Candida krusei (ATCC 6258), lane 7) Aspergillus fumigatus (ATCC 14110), lane 8) Aspergillus flavus (ATCC 64025), lane 9) penicillium chrysogenum (clinical isolate), lane 10) Fusarium solani (clinical isolate), lane 11) Cryptococcus neoformans (ATCC 9011), lane 12) Escherichia coli (ATCC 43894), lane 13) negative control (sterile distilled water), and lane M) a 100-bp molecular size marker; (B) Determine the detection limit of this pan-Candida primers; lane 1) negative control (sterile distilled water), lane2) 2-7 nails treated with serially diluted standard strain (C. albicans (ATCC 5982)) at a concentration of 101-106 CFU/ml, lane 8 positive control (C. parapsilosis (ATCC 4344)) and lane M) a 100-bp molecular size marker
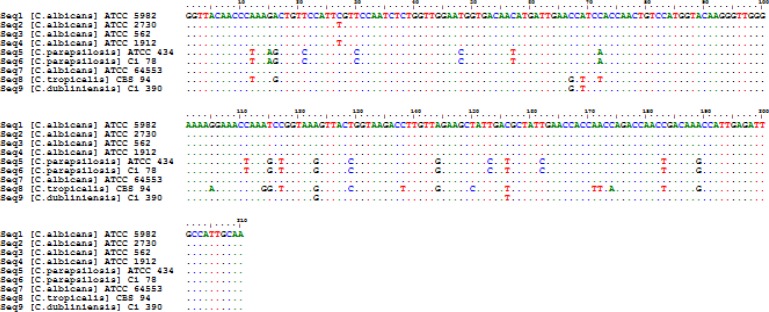
The sequence difference count matrix showed poor intraspecies differences among the species belonging to Candida (0-2 nt insertions/deletions). Pairwise and multiple alignment rates were also evaluated among the sequences. The mean similarity of two sequences for 9 Candida isolates was obtained as 94.6%, and the overall similarity of the sequences was estimated at 86.6%. Figure 2 depicts the multiple DNA sequence alignment of EF-1a sequences in the most common pathogenic Candida tested in this study. The consensus nucleotide sequence data determined in this study were deposited in the GenBank, under the accession numbers of MK568517-MK568521 and MN231082-MN231085.
Figure 2.
Multiple sequence alignments of translation elongation factor 1-alpha gene sequence from various Candida species sequenced in this study
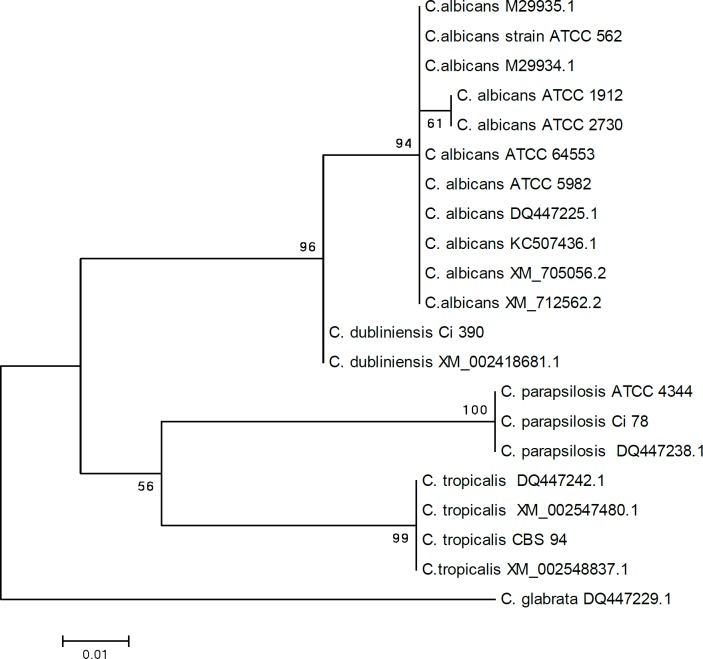
Phylogenetic tree of TEF-1α sequences in Candida species indicated that the nine strains tested in this study (i.e., C. parapsilosis clinical isolate Ci 78, C. tropicalis CBS 94, C. dubliniensis Ci 390, C. albicans ATCC 5982, C. albicans ATCC 2730, C. albicans ATCC 562, C. albicans 1912, C. albicans ATCC 4344, and C. albicans ATCC 64553) were most closely related with strains derived from the gene bank and shared a common branch. In the phylogenetic tree, the studied species were divided into two clades. Clad one consisted of two clusters of C. albicans and C. dublinensis and clad two contained three species of C. parapsilosis, C. tropicalis, and C. glabrata (Figure 3).
Figure 3.
Phylogenetic analysis of elongation factor 1-alpha gene of 9 DNA isolates (standard and clinical isolates) of Candida species tested in this study and a number of sequences in this gene region related to Candida species in the gene bank (NCBI)
For the determination of the analytical detection limit of the designed PCR reaction, serially diluted DNAs were assayed by pan-Candida PCR (Figure 1 right). According to the results, 101 cells/mL was the minimum number for reliable detection.
Out of the 137 tested nail samples suspected of onychomycosis, 35% (n=48), 30.7% (n=42), and 40.1% (n=55) of the specimens were positive for yeast elements by direct microscopy examination, culture, and pan-Candida PCR, respectively (Table 2).
Table 2.
Agreement of microscopy and Pan-Candida PCR methods with the gold standard method (culture)
| Culture | Total | |||
|---|---|---|---|---|
| True positive | True negative | |||
| Microscopy | Positive | n=42 | n=6 | n=48 |
| Negative | n=0 | n=89 | n=89 | |
| Pan- Candida PCR | Positive | n=35 | n=20 | n=55 |
| Negative | n=7 | n=75 | n=82 | |
| Total | n=42 | n=95 | n=137 | |
PCR: polymerase chain reaction
The proportion of patients with positive pan-Candida PCR was higher than that of the patients with positive culture for Candida species (30.7% vs. 40.1%, respectively). This difference was statistically significant (P<0.001). Considering culture as the reference method to assess the performance of the molecular method used in this study, the PPV, NPV, specificity, and sensitivity were calculated as 63.3%, 91.4%, 78.9%, and 83.3% for pan-Candida PCR test, respectively.
Discussion
Rapid and accurate detection of Candida species in nail specimens is essential for the initiation of proper antifungal treatments. Among the routine methods, KOH preparation is easy to perform and economic; however, its sensitivity is inconsistent and influenced by such factors as inadequate sample volume, different sampling methods, and experience of the lab workers. Although fungal culture can identify specific pathogens, it takes a long incubation period and its sensitivity is much lower than that of KOH. In this regard, the false-negative rate of culture for the diagnosis of onychomycosis was approximately 30%, and the sensitivity was about 60% [17].
DNA-based diagnostic tests are increasingly being employed in the clinical microbiological laboratory for the detection of the etiological agent directly from the clinical specimens. The sensitivity of the detection molecular methods depends on various factors, one of the most important of which is the selection of the target DNA for amplification. Over the last two decades, the use of molecular diagnostic study for the detection of a specific sequence of Candida to investigate intra-species and inter-species differences has been reviewed [18, 19]. In such studies, when the main purpose was to design pan-Candida primers, the designed primer pair usually amplified common environmental contaminants, such as Aspergillus. The present study involved the design of a specific pan-Candida primer set in the TEF-1α gene where the primer sequences are conserved within Candida species but distinct from a variety of fungi other than Candida species and the bacterium Escherichia coli. The nucleotide sequence of TEF-1α gene encoding a part of the protein translation machinery appears to be consistently single copy. The only study investigating TEF-1α gene in Candida was conducted in 1990 by Sundstrom et al. to determine whether any of the characteristics of fungal TEF-1α proteins could be used to delineate the phylogenetic relationship of C. albicans with other fungi. Sequence analysis of the study showed the presence of two genes, called TEFI and TEF2, for TEF-1α in C. albicans. The coding regions of TEFI and TEF2 differed by only five nucleotides and encoded identical TEF-1α proteins of 458 amino acids [20].
This study represents the first application of a PCR-based diagnostic test for the most common Candida species using primers amplifying 270-bp region of the TEF-1α gene. Our results showed a great increase in the detection rate of Candida species using pan-Candida PCR (40.01%) over the direct microscopy (35%) and culture (30.7%). In the same vein, Khan et al. obtained more positive results by PCR than conventional methods (52.2% % vs. 21.2 %) [21].
In our study, 7 out of 42 samples which were positive in culture showed negative results in pan-Candida PCR. The DNA from these samples was diluted, and in another reaction, was exposed to pan-fungal primers (ITS1 and ITS4); however, the results stayed negative in the PCR. The positive and negative controls in all experiment runs ruled out the possibility of experimental error. The few negative results can be due to the fact that positive material was not included in the subsample set used for molecular testing.
The pan-Candida PCR detected Candida in 20 samples identified negative by culture. This nonconformity that reduce the specificity (78.9%) and PPV (63.3%) of pan-Candida PCR has been linked rather to false-negative results of the culture method because PCR is more sensitive than culture methods and can detect all pathogenic fungi, both viable nonviable types.
Our pan-Candida primers facilitated the detection of DNA for all tested Candida species, except for C. glabrata. This finding is in the same line with the results obtained by Zhang et al. evaluating four pan-Candida primer sets by conventional PCR assays [22].
The lower limit of detection in this assay was 101 cells/mL which could be sufficient to find the low number of fungal elements causing candidal onychomycosis in a small volume of nail samples although this would require a highly efficient extracted template DNA. Candida DNA was detected in a few nail samples obtained from healthy volunteers, suggesting that this represented commensal fungi living on the nails. Accordingly, such methods as quantitative real-time PCR can be important for the appropriate evaluation of clinical samples containing both pathogenic and commensal fungi.
Conclusion
As the results of the present study indicated, pan-Candida PCR assay showed good performance characteristics by increasing the detection rates of Candida species and drastically reducing the clinical turnaround time, compared to culture. However, it is required to perform further research using more types of standard isolates and larger clinical samples to define the practical value of such an approach in the mycology laboratory.
Acknowledgments
This study was extracted from a master thesis submitted by Forough Farazmand and funded by the Deputy of Research and Technology of Shiraz University of Medical Sciences, Shiraz, Iran (grant No. 18245).
Author’s contribution
M. M. and K. P. supervised all parts of the project and wrote the paper. H. M. designed the study. F. F., F. G. H., R. A., and H. G. set up and performed the tests. M. K. supported DNA sequencing. K. Z. edited the final manuscript.
Conflicts of interest
No potential conflicts of interest have been declared. The authors alone are responsible for the content and writing of the paper.
Financial disclosure
The authors received no external funding for this study.
References
- 1.Xiang H, Xiong L, Liu X, Tu Z. Rapid simultaneous detection and identification of six species Candida using polymerase chain reaction and reverse line hybridization assay. J Microbiol Methods. 2007;69(2):282–7. doi: 10.1016/j.mimet.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Low CY, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep. 2011;3:14. doi: 10.3410/M3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jesudanam TM, Rao GR, Lakshmi DJ, Kumari GR. Onychomycosis: a significant medical problem. Indian J Dermatol Venereol Leprol. 2002;68(6):326–9. [PubMed] [Google Scholar]
- 4.Dubljanin E, Dzamic A, Mitrovic S, Arsenijevic VA, Calovski IC. Onychomycosis: clinical findings, etiological agents and evaluation of laboratory methods. Arch Biol Sci. 2014;66(2):587–94. [Google Scholar]
- 5.Selvarangan R, Bui U, Limaye AP, Cookson BT. Rapid identification of commonly encountered Candida species directly from blood culture bottles. J Clin Microbiol. 2003;41(12):5660–4. doi: 10.1128/JCM.41.12.5660-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillus spp from clinical samples using real‐time PCR. Clin Microbiol Infect. 2006;12(8):745–53. doi: 10.1111/j.1469-0691.2006.01498.x. [DOI] [PubMed] [Google Scholar]
- 7.Taverna CG, Bosco-Borgeat ME, Murisengo OA, Davel G, Boite MC, Cupolillo E, et al. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem Inst Oswaldo Cruz. 2013;108(2):178–85. doi: 10.1590/0074-0276108022013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbedo LS, Figueiredo-Carvalho MH, Muniz Mde M, Zancopé-Oliveira RM. The identification and differentiation of the Candidaparapsilosis complex species by polymerase chain reaction-restriction fragment length polymorphism of the internal transcribed spacer region of the rDNA. Mem Inst Oswaldo Cruz. 2016;111(4):267–70. doi: 10.1590/0074-02760150466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen R, Torp M, Kosiak B, Holst-Jensen A. Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycol Res. 2005;109(Pt 2):173–86. doi: 10.1017/s0953756204002114. [DOI] [PubMed] [Google Scholar]
- 10.Silva FP, Vechiato MH, Harakava R. EF-1α gene and IGS rDNA sequencing of Fusarium oxysporum f sp vasinfectum and F oxysporum f sp phaseoli reveals polyphyletic origin of strains. Trop Plant Pathol. 2014;39(1):64–73. [Google Scholar]
- 11.Voigt K, WoÈstemeyer J. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene. 2001;270(1-2):113–20. doi: 10.1016/s0378-1119(01)00464-4. [DOI] [PubMed] [Google Scholar]
- 12.Mirhendi H, Makimura K, de Hoog GS, Rezaei-Matehkolaei A, Najafzadeh MJ, Umeda Y, et al. Translation elongation factor 1-α gene as a potential taxonomic and identification marker in dermatophytes. Med Mycol. 2014;53(3):215–24. doi: 10.1093/mmy/myu088. [DOI] [PubMed] [Google Scholar]
- 13.Carberry S, Neville CM, Kavanagh KA, Doyle S. Analysis of major intracellular proteins of Aspergillus fumigatus by MALDI mass spectrometry: identification and characterisation of an elongation factor 1B protein with glutathione transferase activity. Biochem Biophys Res Commun. 2006;341(4):1096–104. doi: 10.1016/j.bbrc.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 14.Ignjatovic VA, Stevanovic MT, Durdevic VS, Petrovic MM, Stanucevic MP, Dzamic AM, et al. Onychomycosis: sampling, diagnosing as efficiant part of hospital pharmacology. Hosp Pharmacol Int Multidisciplinary J. 2014;1(3):130–7. [Google Scholar]
- 15.Abd-Elsalam KA. Bioinformatic tools and guideline for PCR primer design. Afr J Biotechnol. 2003;2(5):91–5. [Google Scholar]
- 16.Silva GA, Bernardi TL, Schaker PD, Menegotto M, Valente P. Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz Arch Biol Technol. 2012;55(2):319–27. [Google Scholar]
- 17.Petinataud D, Berger S, Ferdynus C, Debourgogne A, Contet‐Audonneau N, Machouart M. Optimising the diagnostic strategy for onychomycosis from sample collection to fungal identification evaluation of a diagnostic kit for real‐time PCR. Mycoses. 2016;59(5):304–11. doi: 10.1111/myc.12471. [DOI] [PubMed] [Google Scholar]
- 18.Barbedo LS, Figueiredo-Carvalho MH, Muniz MN, Zancope-Oliveira RM. Comparison of four molecular approaches to identify Candidaparapsilosis complex species. Mem Inst Oswaldo Cruz. 2017;112(3):214–9. doi: 10.1590/0074-02760160412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanbe T, Horii T, Arishima T, Ozeki M, Kikuchi A. PCR‐based identification of pathogenic Candida species using primer mixes specific to Candida DNA topoisomerase II genes. Yeast. 2002;19(11):973–89. doi: 10.1002/yea.892. [DOI] [PubMed] [Google Scholar]
- 20.Sundstrom P, Smith D, Sypherd PS. Sequence analysis and expression of the two genes for elongation factor 1 alpha from the dimorphic yeast Candidaalbicans. J Bacteriol. 1990;172(4):2036–45. doi: 10.1128/jb.172.4.2036-2045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan ZU, Mustafa AS. Detection of Candida species by polymerase chain reaction (PCR) in blood samples of experimentally infected mice and patients with suspected candidemia. Microbiol Insights. 2001;156(1):95–102. doi: 10.1078/0944-5013-00072. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Hung GC, Nagamine K, Li B, Tsai S, Lo SC. Development of Candida-specific real-time PCR assays for the detection and identification of eight medically important Candida species. Microbiol Insights. 2016;9:21–8. doi: 10.4137/MBI.S38517. [DOI] [PMC free article] [PubMed] [Google Scholar]