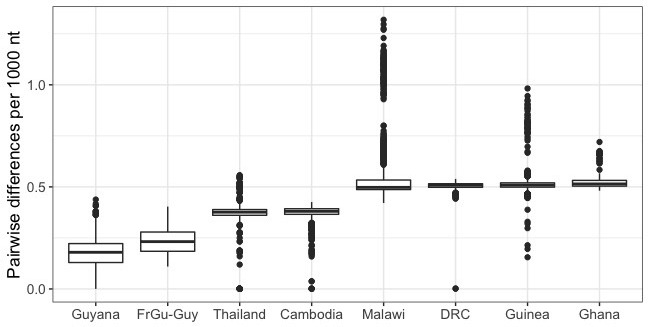
Figure 2. Whole-genome sequence analysis of pfk13 C580Y mutant parasites in Guyana.
(a) Comparison of the haplotypic background of pfk13 C580Y mutant parasites from Guyana, 2016, and Southeast Asia, 2010–2012. Across Pf3k samples from Cambodia, Thailand, and Vietnam, 45 unique C580Y-coding haplotypic backgrounds were identified and compared to haplotypes from Guyana. Columns represent 149 sites containing non-singleton single nucleotide polymorphisms (SNPs) found within a 150 kb segment surrounding the pfk13 C580Y-coding allele. At a given site, the more common allele is marked blue, the less common allele is orange, and missing calls are grey. The Y-coding variant for codon 580 of pfk13 is represented by the red blocks; wild-type is blue. Only the five pfk13 C580Y mutant samples with fewer than 15% missing calls are depicted here. (b) Analysis of relatedness at the whole-genome level among Guyana clones. Pairwise identity-by-descent (IBD) was estimated for all pairs of Guyana samples with high quality whole-genome sequence data (<70% missing calls). Pairwise comparisons between samples exhibiting the pfk13 C580Y allele are indicated in red, and show uniformly high levels of relatedness, suggesting a single clonal lineage harboring the resistance mutation. (c, d) Principal components analysis of parasites from Guyana or other geographic regions using SNP calls from whole-genome sequence data. (c) The parasites from Guyana and French Guiana form a single cluster when compared with parasites from Africa. (d) The two edited parasite lines from French Guiana are highly similar to the sequenced parasite samples from Guyana including a pfk13 mutant.
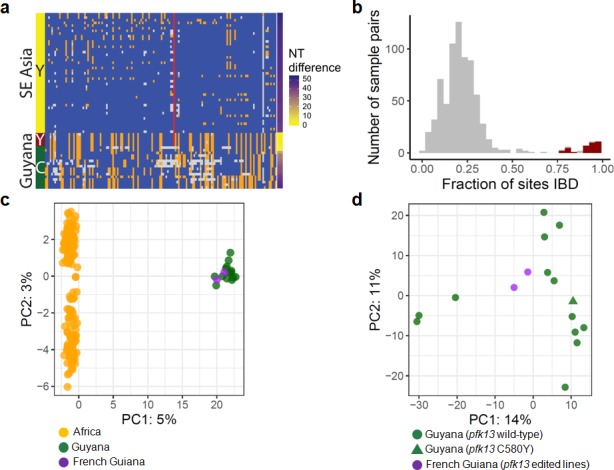
Figure 2—figure supplement 1. Number of single nucleotide differences between pairs of parasites within different geographic locations.