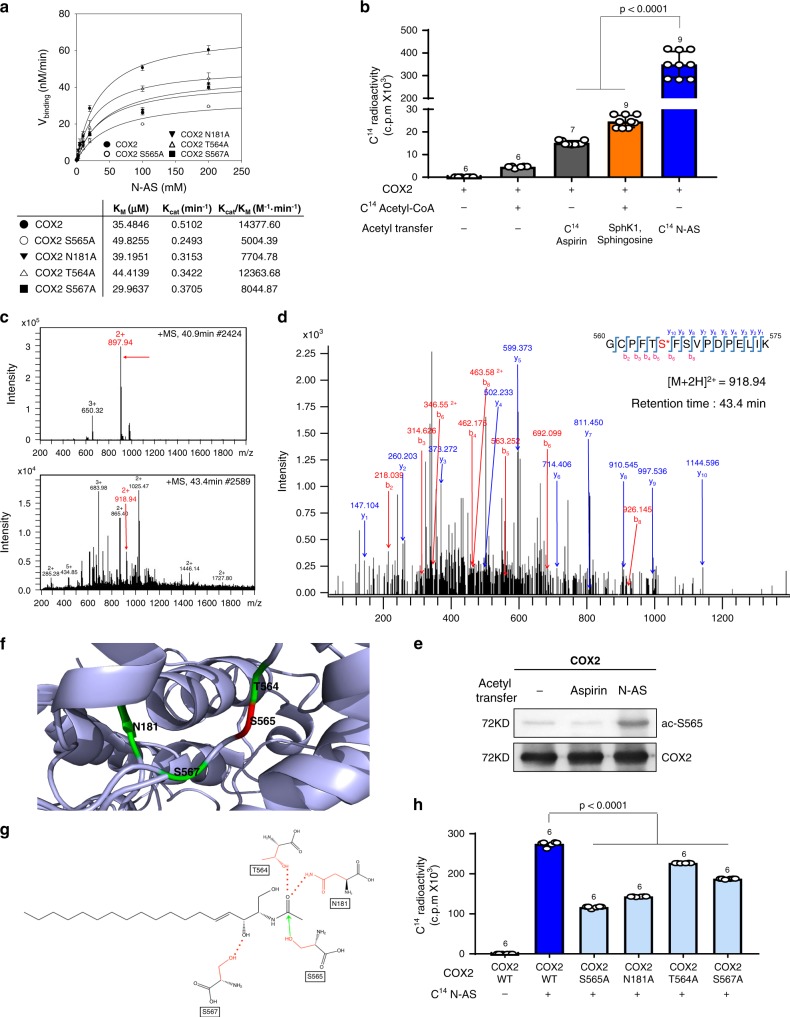
Fig. 1. N-AS acetylates S565 in COX2.
a N-AS binding activity of COX2 WT, COX2 S565A, COX2 N181A, COX2 T564A, and COX2 S567A was analyzed by filter binding assay. The binding velocity (Vbinding) of [14C] N-AS to COX2 was plotted to the N-AS concentration and the nonlinear regression analysis of the saturated plot yielded the kinetic parameters such as Kcat (catalytic constant) and KM (Michaelis–Menten constant) for N-AS and COX2 binding activity (n = 3 independent experiments per group). b Acetylation assay of purified COX2 protein treated with [14C] N-AS. The purified COX2 protein incubated in the presence of [14C] N-AS for 2 h at 37 °C and then COX2 was analyzed on scintillation counter. [14C] aspirin-treated COX2 protein and [14C] acetyl-CoA treated COX2 protein with SphK1 and sphingosine were positive control (n = 6–9 per group). c LC-MS spectra of peptide 560-GCPFTSFSVPDPELIK-575 (m/z = 918.94) of COX2 acetylated by N-AS. d LC-MS/MS spectra of ac-S565 in 560-GCPFTSFSVPDPELIK-575 of COX2. e Recombinant COX2 protein was incubated in the presence or absence of 2 mM N-AS or aspirin as indicated and S565 acetylation was determined by western blotting using anti-ac-S565 and COX2 antibodies. Data were replicated in six independent experiments with similar results. f The theoretical crystallographic model of human COX2 structure was taken from Protein Data Bank file 1V0X. S565 and residues (N181, T564, and S567) involved in the catalysis of COX2 were projected on the crystal structure of COX2 using PyMOL. Residues are shown as well: (1) S565 (red); (2) N181, T564, and S567 (green). g Proposed mechanism for the acetylation of S565 of COX2 by N-AS. h Acetylation assay of COX2 WT, COX2 S565A, COX2 N181A, COX2 T564A, and COX2 S567A treated with [14C] N-AS. (n = 6 independent experiments per group). b, h One-way analysis of variance, Tukey’s post hoc test. All error bars indicate s.e.m. Source data are provided as a Source data file.