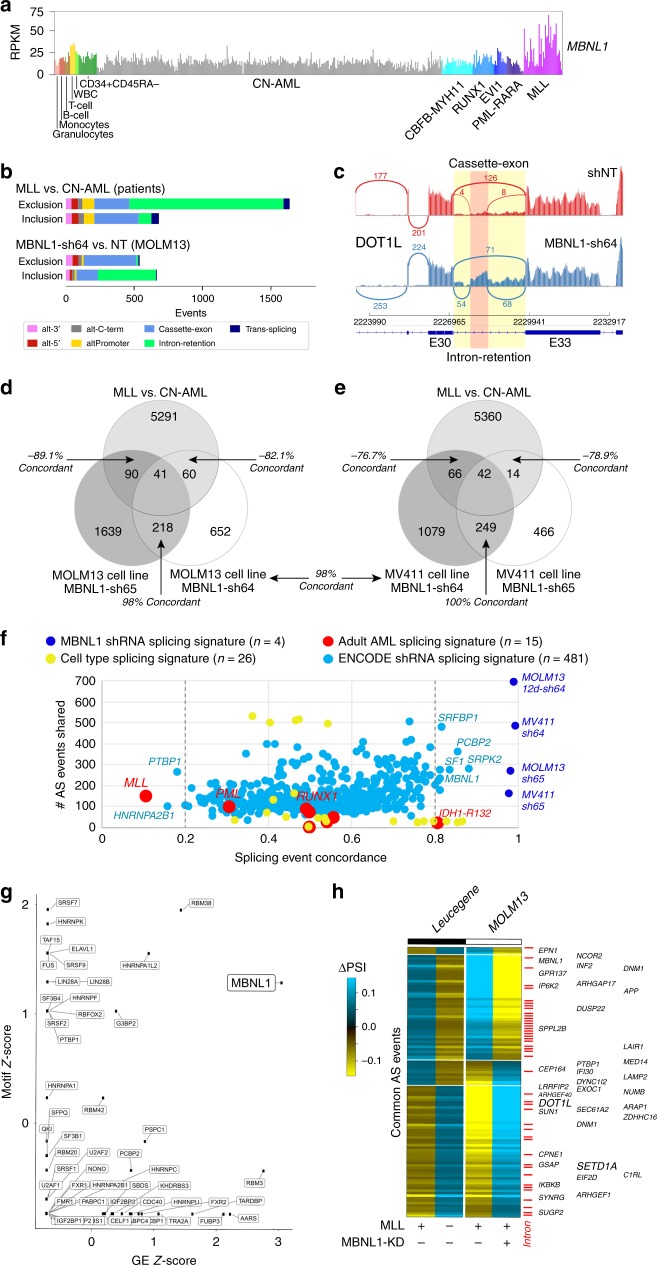
Fig. 6. Partial reversal of impaired intron retention in MLL-rearranged cells by MBNL1 knockdown.
a MBNL1 gene expression levels in adult patient AML samples according to genetically/cytogenetically defined subtypes and for sorted cell populations. b Breakdown of the type of alternative splicing events detected when comparing Leucegene AML diagnostic patient RNA-Seq samples with prior reported MLL oncofusions versus cytogenetically normal patients (CN-AML), or events obtained from MOLM13 AML cells bearing the MLL-AF9 karyotype with transfection of a short-hairpin RNA for MBNL1 (sh64) versus shRNA control (NT). Splicing-events that demonstrate increased exon inclusion versus exon-exclusion are shown separately. c SashimiPlot illustrating increased intron retention and cassette splicing of the MLL interacting protein DOT1L, resulting in alternate, likely truncated isoform in MBNL1 knockdown cells (shRNA construct 64). Exon number definitions are based on AltAnalyze Ensembl 72 definitions. d, e Overlapping alternative splicing events from MLL oncofusion patient samples compared to those from two different MLL-rearranged cell lines (MOLM13 (d), MV4;11 (e)), using two separate short hairpin RNAs against MBNL1 (sh64, sh65). Callouts highlight the number of splicing events occurring anti-concordantly between the Leucegene MLL vs. CN-AML and shMBNL1 vs shNT comparison, or AS events occurring in the same direction (shMBNL1-64 and -65 overlap). Concordance percentages are indicated next to each overlap. Concordance between MV4;11 (sh64) and MOLM13 (sh65) is indicated between the two Venn diagrams. f Unbiased comparison of splicing signatures associated with different AML genetic/cytogenetic subtypes, prior splicing factor knockdown signatures and hematopoietic cell types relative to that of MBNL1 knockdown. Splicing signatures were derived using the same pipeline for determining MBNL1 knockdown and MLL rearrangement-associated splicing events. Similarity to the MBNL1 knockdown splicing signature (specifically shMBNL1-64 in MOLM13) is shown in terms of splicing concordance, similar to (d) and (e), indicating the number of splicing events common to each signature on the Y-axis. Fifty-percent concordance is representative of random overlap. For each group, n represents number of pairwise comparisons (i.e., AML with fusion/molecular alteration vs CN-AML for Leucegene, RBP knockdown vs non-targeting control for ENCODE, hematopoietic cell type vs stem/progenitor, or shMBNL1 vs non-targeting control. g Statistical enrichment (z-score) of defined RNA-binding protein RNA-recognition elements (motifs) from the CisBP-RNA database for alternatively splicing exonic and intronic intervals with MBNL1 knockdown (sh64) compared to enrichment of differential gene expression (z-score). h Heatmap of all overlapping alternative splicing events in Leucegene MLL versus CN-AML and those observed with knockdown of MBNL1 (sh64) in MOLM13 cells, with callout of genes with prior implicated cancer relevance (ToppGene database). Red ticks indicates intron-retention events predicted from MultiPath-PSI.