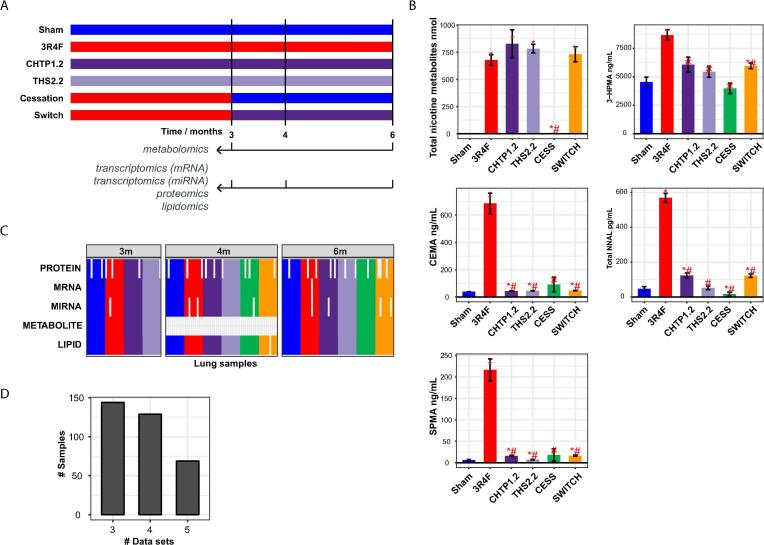
Fig. 1.
Multi-omics analysis of lung exposure effects. (A) Design of a 6-month inhalation study on ApoE−/− mice. Other endpoints and additional details on this systems toxicology study are reported in Phillips et al. [23]. (B) Exposure markers measured in urine: 3-hydroxypropylmercapturic acid (3-HPMA), a biomarker of acrolein uptake; 2-cyanoethylmercapturic acid (CEMA), a biomarker of acrylonitrile uptake; total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), a biomarker of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone uptake; and S-phenylmercapturic acid (SPMA), a biomarker of benzene uptake. Note that the high basal levels of 3-HPMA, including in the Sham group, can be explained by endogenous acrolein production [71]. Data are shown as mean ± standard error of the mean. Statistical comparisons: *p value versus Sham <0.05; #p value versus 3R4F <0.05; N = 10–12. (C) Lung samples obtained and analyzed from the same animals across the five omics analyses. A colored cell in the matrix indicates that a lung sample from an animal was analyzed successfully for the given omics dataset. Colors of sample groups as in panel B. Analyses performed with a planned N = 9 for mRNA/miRNA transcriptomics, lipidomics, and metabolomics and with a planned N = 8 for proteomics. Four-month samples were not submitted for metabolomics analysis. (D) Distribution plot showing the number of shared samples across a given number of omics datasets.