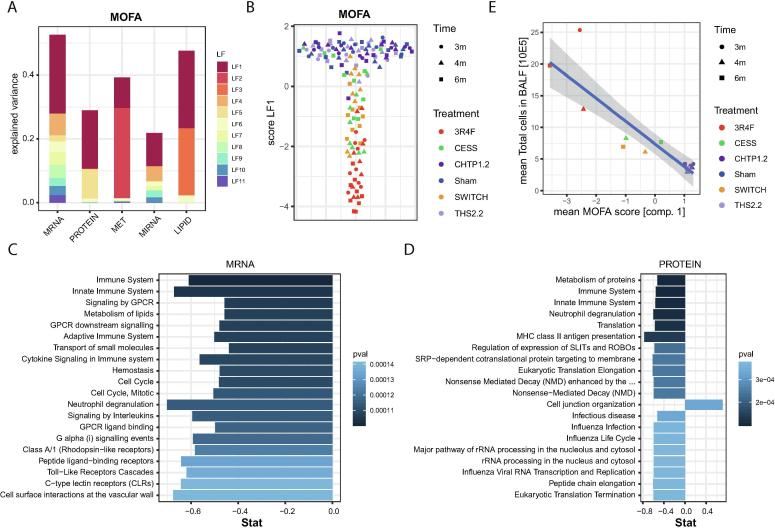
Fig. 3.
Integrative multi-omics LF model. An integrative LF model for all five data modalities was derived by MOFA [50]. (A) Fraction of explained variance for each data modality and each discovered LF of the MOFA model. (B) Score plot for LF 1. The LF 1 score for each sample is given on the y-axis. (Random spread on x-axis for visualization purposes.) The shape of the data points indicates the time point, and the colors indicate the treatment or exposure times (see key). (C, D) Top 20 enriched gene sets (by p value) for the weight vectors of LF 1 for the mRNA transcriptomics (C) and proteomics (D) datasets. (E) Comparison between the mean MOFA score of LF 1 and the total number of (immune) cells in BALF for each treatment and time-point group (see key). Linear fit with the 95% confidence interval band is shown (blue line, grey area). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)