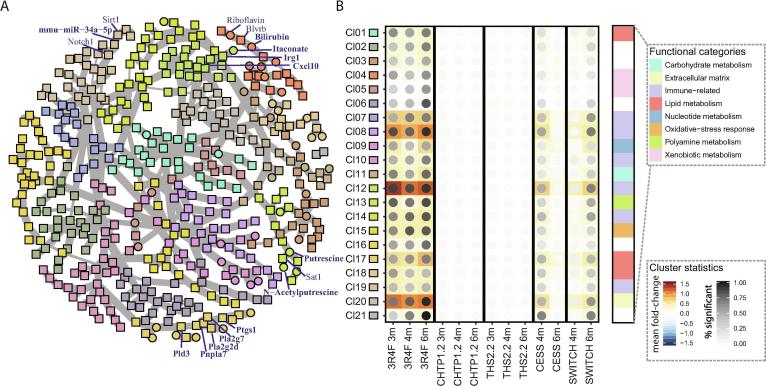
Fig. 4.
Multi-omics response network. (A) Aggregated gene–protein–metabolite–miRNA network for LF 1 constructed by using the PCSF algorithm [55]. Nodes represent molecules, and edges represent their interactions. Node colors reflect the identified clusters/communities in the network (see cluster key in panel B). Select molecules are labeled. See the interactive graph in Supplementary Fig. 5 for details [Supplementary File 1]. (B) Expression profiles for identified clusters (Cl). Mean log2 fold changes are color-coded, and the percentage of molecules with significant differential expression/abundance in a cluster is indicated by the darkness of the dots (see key). In addition, the broad functional classifications for the clusters are indicated (color bar on right side).