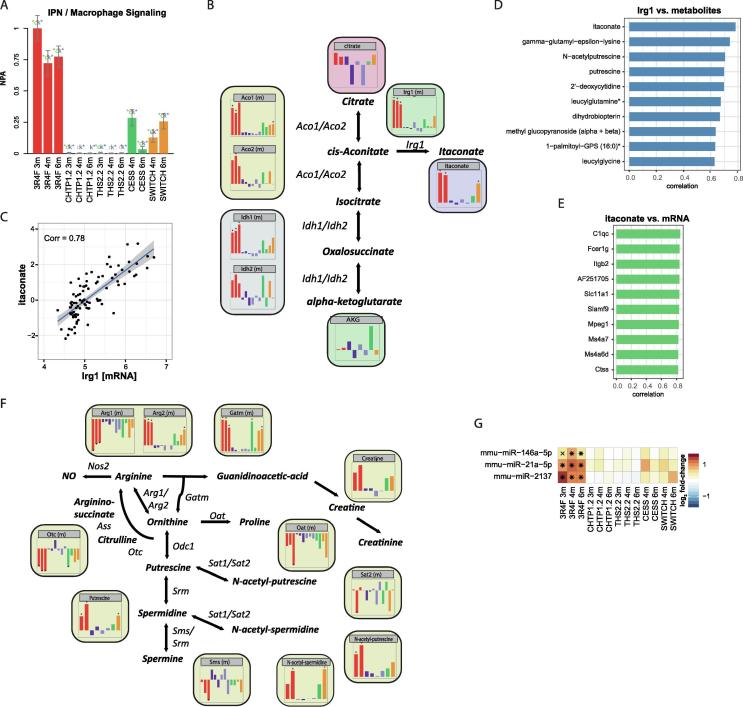
Fig. 5.
Metabolite and miRNA changes associated with 3R4F CS-induced immune response. (A) Network enrichment analysis of the macrophage signaling network. The bars show the overall NPA based on transcriptomics data; error bars show the 95% confidence interval. Three statistical measures are shown: the red star indicates statistical significance with respect to biological replicates; the green star (o statistic) indicates significance with respect to permutation of genes downstream of the network nodes; and the blue star (k statistic) indicates significance with respect to permutation of the network topology (p < 0.05). (B) Itaconate metabolic pathway. Bar charts show log2 fold-change responses versus Sham, ordered and colored as in panel A. (C) Correlation between Irg1 mRNA expression and itaconate abundance. Blue line shows fit from the linear model, with the 95% confidence interval band in grey. The correlation coefficient (Corr) is indicated. (D) Top 10 correlations of Irg1 mRNA expression against all metabolites. (E) Top 10 correlations of itaconate abundance against all mRNAs. (F) Polyamine pathway, as in panel B. (G) Expression profiles for selected immune-related miRNAs. Log2 fold-changes versus Sham are color-coded, and statistical significance is indicated: *FDR-adjusted p value <0.01; XFDR-adjusted p value <0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)