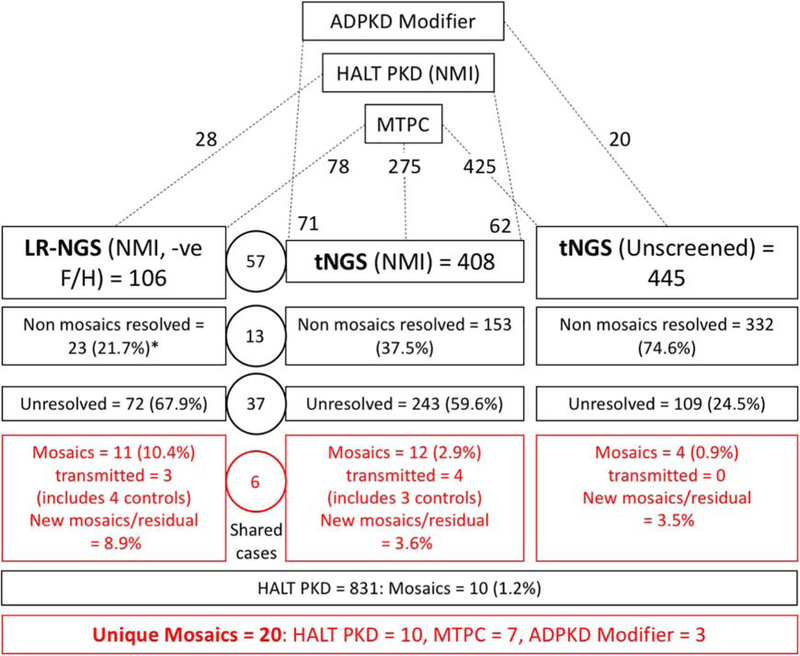
Figure 1. Diagram showing the study design and detection rates for the different groups involved in the screen for mosaicism.
ADPKD patients were included in the study from those collected for mutation screening at the Mayo Translational PKD Center (MTPC), ADPKD Modifier patients, and PKD1 and PKD2 SS NMI HALT PKD patients. The analysis for mosaic cases was performed in three screens, LR-NGS of PKD1 and PKD2 SS NMI ADPKD patients with a proven or suspected negative family history (-ve F/H), a tNGS screen of PKD1 and PKD2 SS NMI ADPKD patients, and a tNGS screen of previously unscreened patients. The number of total families included in each screen, and their origin is indicated at the top portion of the figure. The detection rate breakdown of each screen is indicated in boxes and includes number of families resolved with typical heterozygous variants (non mosaics resolved), number of families remaining unresolved, and the number of families with detected mosaic variants. The numbers in the circles represents families shared among the LR-NGS and tNGS approach. The number of newly detected mosaics as a proportion of the unresolved plus mosaics (New mosaics/residual) is also calculated. Approximately 1% of not families (HALT PKD (1.2%) and tNGS (Unscreened; 0.9%)) were mosaics. In addition, newly resolved mosaics represented 8.9% of the unresolved plus mosaic families from the LR-NGS screen (-ve F/H and SS PKD1/PKD2 NMI) and 3.6% from the tNGS (SS PKD1/PKD2 NMI) screen. The number of unresolved families from the tNGS (unscreened) analysis is greater than expected from a typical ADPKD population, which is likely because of the large number of very mild and atypical patient screened from the MTPC population. *For the non-mosaic patients resolved from the LR-NGS screen, results from other genes detected by tNGS are also included.