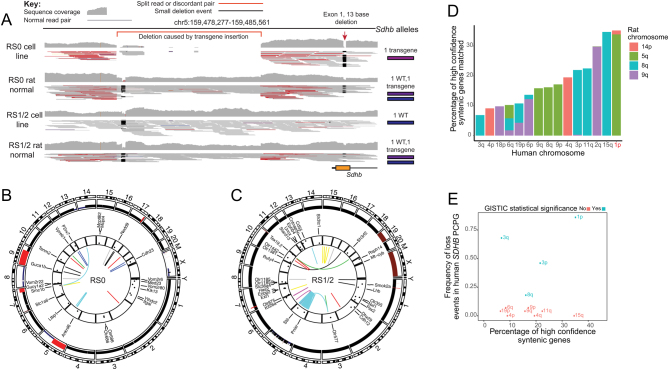
Figure 6.
WGS of RS0 and RS1/2 tumor cell lines and their matched germline DNA. (A) Genome viewer snapshot of sequence coverage spanning exon 1 of Sdhb and the genomic region immediately 5′ of Sdhb (~1.4 kb in total). A 13-bp deletion within exon 1 of Sdhb was detected in the germline DNA of both RS0 and RS1/2. The exon 1 deletion was present in all sequencing reads in tumor cell line RS0, indicating that the WT Sdhb allele had been lost during tumorigenesis. Conversely, the TALEN-edited Sdhb allele was lost during tumorigenesis of tumour cell line RS1/2. In addition to the exon 1 deletion, a larger 729-bp deletion was also detected upstream of Sdhb, indicated by the presence of discordant or split reads spanning the deletion breakpoint (these reads are highlighted in red). The deletion upstream of Sdhb is also supported by a precipitous drop in sequence coverage spanning the 729-bp deletion. Like the 13-bp exon 1 deletion, the 729-bp deletion was detectable in the germline DNA of RS0 and RS1/2, indicating that it was introduced by TALEN gene-editing. (B) A circos plot describing the somatic alterations that occurred in the formation of RS0. From outside to inside, tracks are as follows: rat cytoband, total copy-number variation (CNV) as called by FACETS (black signifies n = 2, red signifies n > 2 and blue signifies n < 2), names of genes called by BCBio ensemble variant calling as having a coding mutation, the allele frequency of these mutations and the innermost track represents the structural variants called by GRIDSS. (C) A circos plot describing the somatic alterations that occurred in the formation of RS1/2. Tracks are in the same order as described in (B). (D) The percentage of high confidence orthologous genes matched between rat and human comparing chromosome arms called by FACETS as being altered in RS0 (either gain or loss). (E) Comparison of high confidence orthologous genes from D (further filtered only to chromosomal loss events in RS0) against the frequency of chromosomal arm level loss events in human SDHB PCPG as determined by GISTIC. Human copy-number variation (CNV) data were taken from publicly available copy-number profiles cited in the Materials and methods section. Chromosome arms colored in blue were called as statistically significantly altered by GISTIC (false discovery rate <0.05). Chromosome arm level losses of 1p, 3q, 3p and 8q are frequent and statistically significant events in human PCPG. These regions are overlapping with syntenic genomic regions that have undergone somatic loss in rat model RS0.

 This work is licensed under a
This work is licensed under a