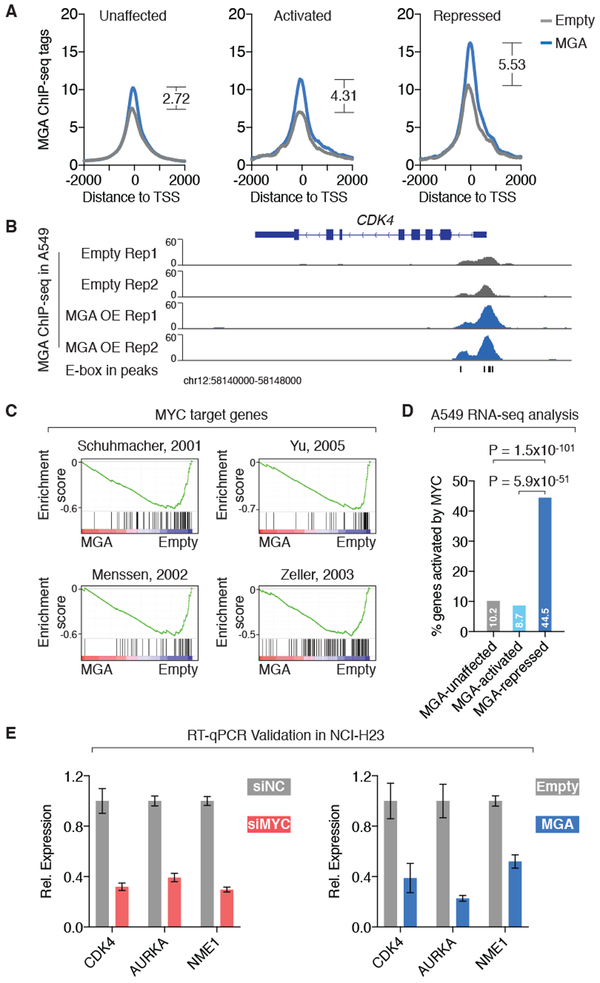
Figure 3: ChIP-seq/RNA sequencing analyses show that MGA binds to and represses MYC target genes.
A. Averaged ChIP-seq signal of MGA in A549 cells with and without MGA overexpression at the transcription start sites (TSS) of genes that are activated, repressed and unaffected by MGA (based on RNA sequencing results in A549 cells with and without MGA overexpression).
B. ChIP-seq signal of MGA in A549 cells with and without MGA overexpression at the CDK4 locus.
C. Gene set enrichment analysis of RNA sequencing results in A549 cells with and without MGA overexpression showed that MGA-repressed genes are enriched in MYC target genes identified by previous studies.
D. Percentage of MGA-regulated genes that are activated by MYC (based on RNA sequencing results from A549 cells with and without siRNA-mediated MYC silencing). P values are derived from fisher exact tests.
E. RT-qPCR assays in NCI-H23 cells validated that either silencing MYC or overexpressing MGA decreased the expression level of the MYC target genes CDK4, AURKA and NME1. Expression level is normalized to controls (siNC or Empty). Error bars: s.d.