Figure 4. NEAT1 modulates neuronal H3K9me2.
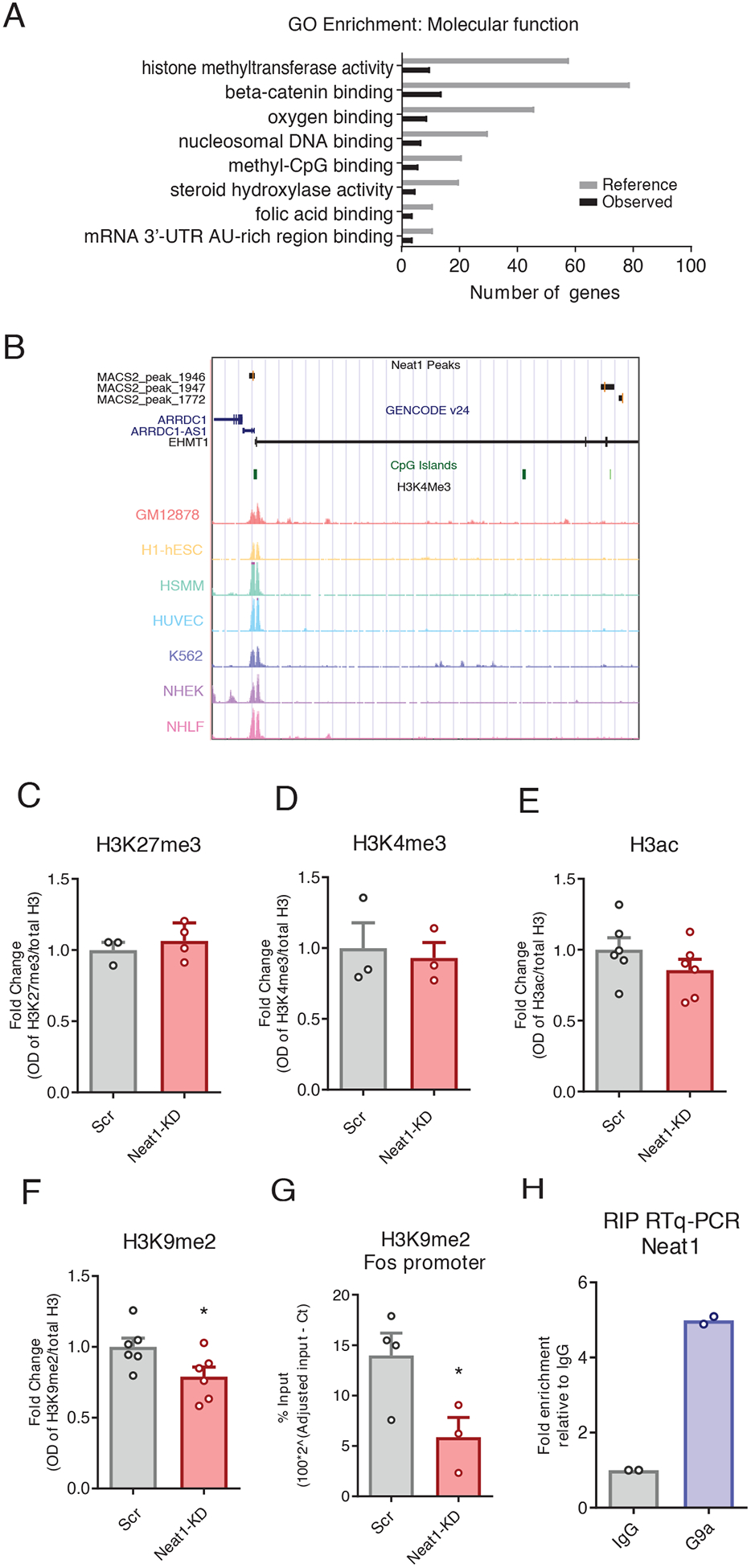
(A) NEAT1 CHART-seq peaks were mapped to the nearest gene transcription start site, and functional enrichment was assessed using ChIP-ENRICH, with histone methyltransferase activity being noted as a significantly enriched GO term (BH corrected, *P < 0.05 for all terms shown). (B) UCSC genome browser plot showing NEAT1-binding peaks overlapping the human EHMT1 gene. (C to G) Analysis of Western blots assessing histone modifications H3K27me3 (C), H3K4me3 (D), H3ac (E), and H3K9me2 (F) globally, and ChIP-qPCR assays at the H3K9me2 at the c-Fos gene promoter (G) in N2a cells after siRNA-mediated knockdown of NEAT1. Data are cultured cells from at least 3 independent experiments, *P< 0.05 by Student’s t-test (F, P=0.0456; G, P=0.0472. (H) RNA-binding protein immunoprecipitation followed by RT-qPCR to assess for Ehmt2/Neat1 interaction in murine N2a cells. Data are means of 2 independent experiments, each represented by a data point.
