Figure 5. PoKI-Seq combines pooled knock-in screening with single-cell RNA sequencing.
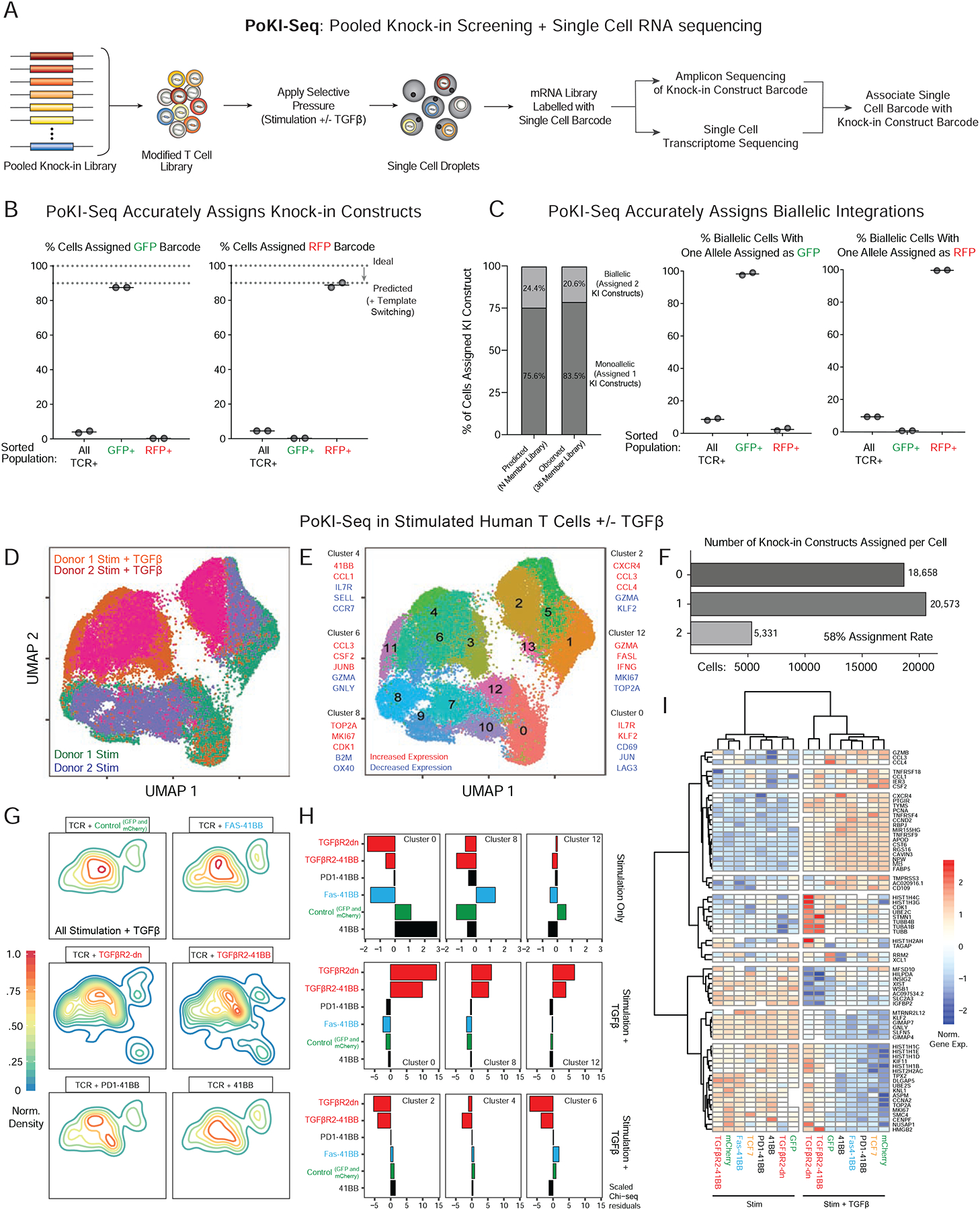
(A) Design of pooled knock-in experiments paired with single cell RNA-sequencing, termed PoKI-seq. This platform provides high-dimensional assessment of cell phenotypes caused by each knock-in construct’s genotype (Figure S6A).
(B) To validate the molecular assignment of knock-in construct barcodes to each individual cell, bulk knock-in positive cells expressing the integrated NY-ESO-1 TCR were sorted, as were NY-ESO-1+ cells that also expressed either GFP+ or RFP+. In the sorted GFP+ and RFP+ populations, the vast majority of template barcodes corresponded to the expression of the expected protein product (Figure 1E).
(C) PoKI-seq identified cells with multiple integrations. The frequency of observed cells with multiple knock-in barcodes likely to due to biallelic integrations closely matched those predicted based on 2-member GFP/RFP library knock-in experiments (Figure 1D). As expected, in sorted GFP+ or RFP+ cells with biallelic integrations, one of the barcodes corresponded to GFP or RFP respectively.
(D) UMAP representation of all single cell states identified in vitro with pooled knock-in T cell populations. 7 days following pooled knock-in of a 35-member library (all constructs in Figure S3A except tNGFR), sorted knock-in positive T cells (NY-ESO-1 TCR+) were stimulated at a 1:1 ratio with CD3/CD28 beads in the presence or absence of exogenous TGFβ and PoKI-seq was performed 4 days later.
(E) Nearest neighbor clustering (Louvain) overlaid on the UMAP representation revealed single cell populations corresponding to distinct cell states and associated hallmark genes.
(F) Assignment of knock-in constructs for each single cell in D. Over 58% of cells were assigned a knock-in construct. Approximately 3.4% of cells were assigned 3 or greater knock-in barcodes, potentially due to sequencing cell doublets, rare imperfect integration of multiple templates or template switching.
(G) Density plots (in the UMAP representation of single cell states) for cells with indicated knock-in constructs in TGFβ-treated conditions.
(H) Over-representation analysis for cells with select knock-in constructs in defined single cell clusters as measured by observed vs. expected frequencies (Chi-square residuals). In the context of stimulation only (top row), the FAS-41BB construct enriched in the proliferative cluster 8. With the addition of exogenous suppressive cytokine TGFβ, cells with TGFβR2-derived knock-in constructs showed strong enrichment in clusters corresponding to proliferative (cluster 8) and effector states (cluster 12), and depletion from the clusters associated with response to TGFβ (clusters 2, 4, 6).
(I) Gene expression heatmap for select knock-in constructs in PoKI-seq experiment. Gene list was generated from genes in the clusters examined in H with absolute log fold change of >0.8 compared to all other clusters. Transcriptional effects of TGFβR2-derived constructs strongly correlated with each other in the presence of exogenous TGFβ but not in the stimulation-only condition. TGFβR2-derived constructs altered the transcriptional response to TGFβ, maintaining expression of genes otherwise associated with the stimulation-only condition, such as proliferative markers MKI67 and TOP2A.
Experiments performed in n=2 (B–I) independent human donors.
See also Figure S6.
