Figure 4. Structural features of JDPs.
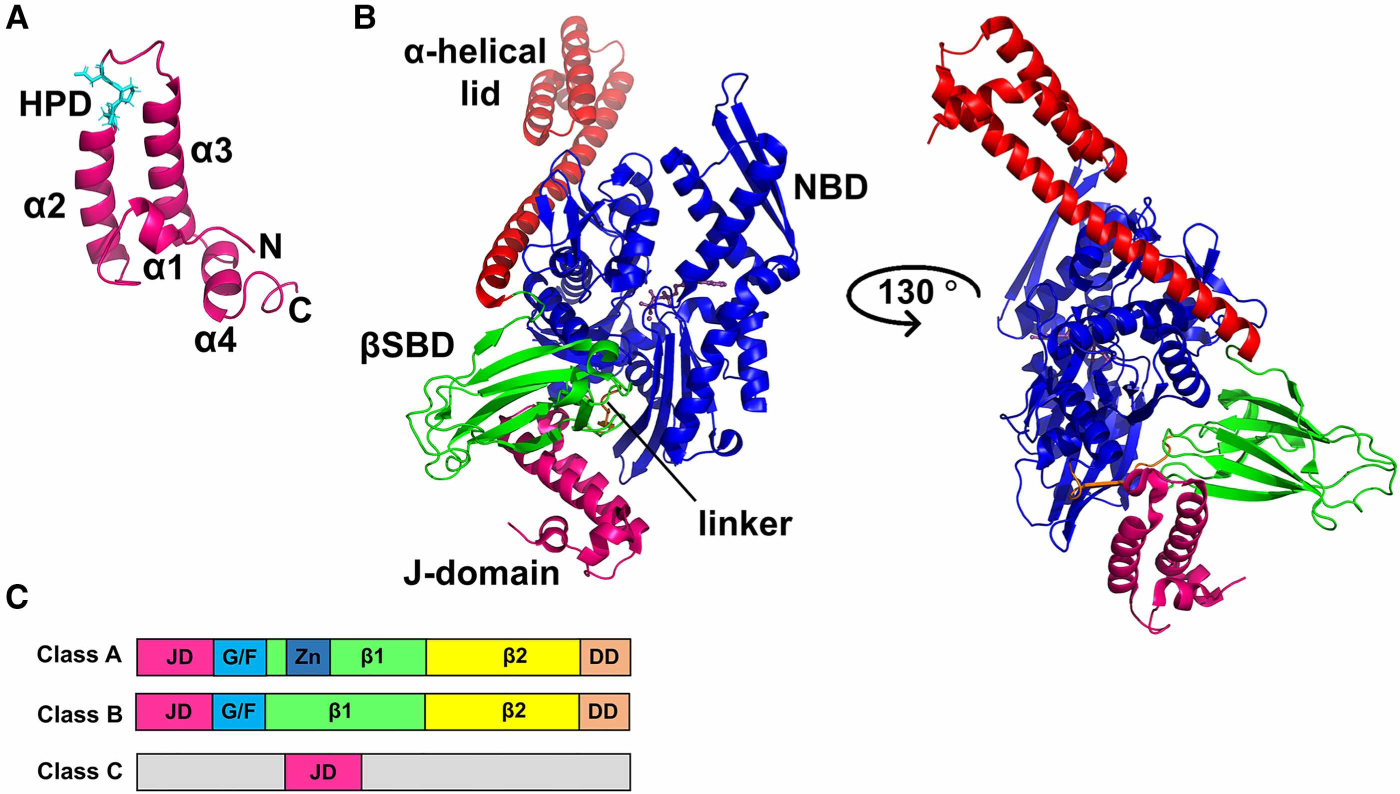
(A) NMR structure of the J-domain of E. coli DnaJ (PDB ID 1xbl [44]) indicating the four helices (α1, helix I; α2, helix II; α3, helix III; α4, helix IV) of the helical hairpin (in purple) and the His–Pro–Asp (HPD) motif (in cyan). (B) Structure of the ATP-bound open conformation of E. coli DnaK in complex with the J-domain of E. coli DnaJ (PDB ID 5nro [36]). The color scheme is the same as that for DnaK in Figure 1, and the J-domain is shown in magenta. The two orientations show that the J-domain binds at the interface between the NBD and the SBD of DnaK, forming direct contacts with both domains. (C) Domain organization and classification of J-proteins; JD, J-domain; G/F, Gly–Phe-rich region; Zn, Zn-finger; β1, first β-sandwich domain; β2, second β-sandwich domain; DD, dimerization domain. In class C JDPs, the functional J-domain can be present anywhere in the sequence and is not restricted to the N-terminus.
