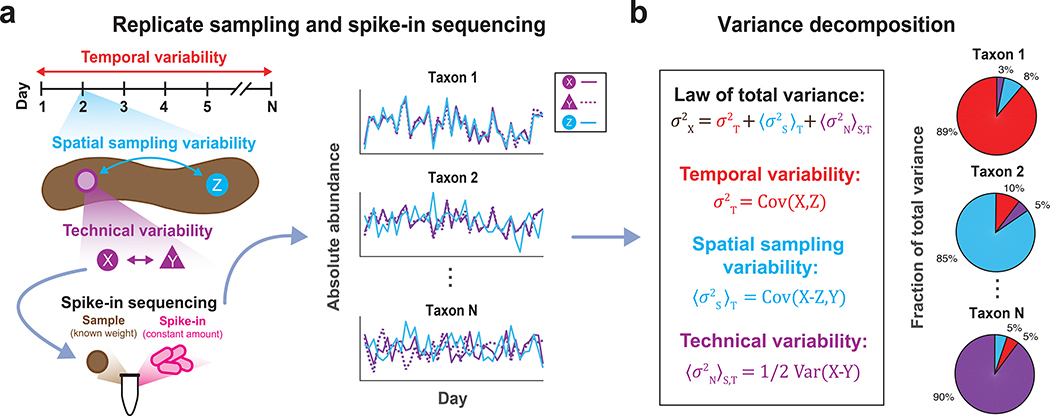
Figure 1 |. DIVERS conceptual workflow.
(a) Illustration of the DIVERS workflow applied to the human fecal microbiome. Samples are collected from two random spatial locations (X and Y from the purple site, Z from the blue site, as shown on the left side of the figure) on each day of sampling and two technical replicates (X and Y) are prepared from one of these spatial locations. The resulting three samples (X, Y, and Z) are subjected to a custom spike-in procedure to estimate absolute bacterial abundances. (b) The DIVERS variance decomposition model is then applied to abundance profiles of each taxa to quantify contributions of temporal variability, spatial sampling heterogeneity and technical noise to total abundance variability.