Fig. 3. LEDGF/p75 suppresses proviral transcription during HIV latency by recruiting PAF1 to maintain RNA Pol II pausing at the provirus.
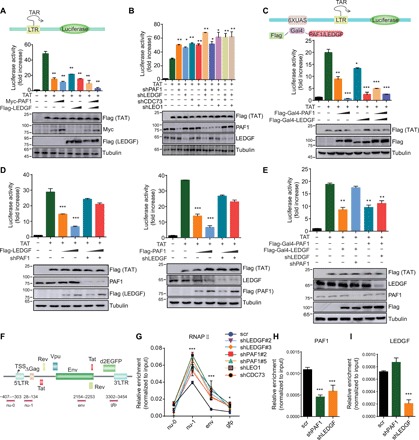
(A) LEDGF/p75 and PAF1 repress Tat-dependent activation of HIV transcription. 293 T cells were transfected with the indicated constructs and the luciferase activity was shown as relative to that in cells only transfected with reporter plasmid (means + SD; n = 3; arbitrarily set to 1). (B) Knockdown of LEDGF/p75 or PAF1 complex elevates the Tat-induced transcriptional activity of the HIV LTR. Luciferase activity in each cellular extract was measured and shown similarly as in (A). (C) Targeting LEDGF/p75 and PAF1 by Gal4-UAS (upstream activation sequence) system suppresses the Tat-induced transcriptional activity of the HIV LTR. (D) Interdependence between LEDGF/p75 and PAF1 on suppression of the Tat-induced transcriptional activity of the HIV LTR. (E) Luciferase reporter assay to examine the activity of Gal4-fused PAF1 or LEDGF/p75 on HIV LTR upon depletion of endogenous LEDGF/p75 or PAF1. (F) Schematic illustration for the location of primers used for the ChIP-qPCR analyses. TSS, transcription start site. (G) ChIP-qPCR analysis of Pol II distribution in E4 cells depleted for the indicated protein for 9 days. Pol II enrichment was normalized to input and shown as means ± SD (n = 3). RNAP, RNA polymerase. (H) PAF1 enrichment at the nu-1 region was determined by ChIP-qPCR, normalized to input, and shown as means + SD (n = 3). (I) LEDGF/p75 enrichment at the nu-1 region was normalized to input and shown as means + SD (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
