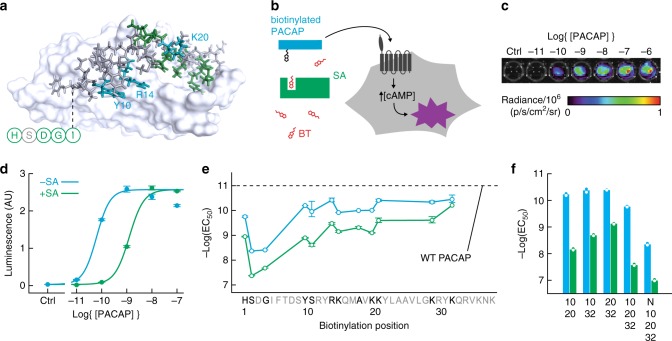
Fig. 2. Engineering a PACAP-based AVATar.
a Identification of candidate ligand attachment sites on PACAP: Structure of PACAP in complex with a PAC1 receptor fragment18, highlighting residues whose substitution blocks (green) or does not affect (blue, labels) peptide bioactivity, according to previous structure-activity studies. N-terminal residues absent from the structure indicated schematically (circles). b Schematic of PAC1-based bioassay for testing biotinylated PACAP constructs (blue), their blocking by streptavidin (green), and release by free biotin (red). Unblocked PACAP derivatives can bind the PAC1 receptor (dark gray) expressed in CHO cells, triggering cAMP production and activating a luminescence reporter (purple). c Raw luminescence recorded from the assay cells under titration with wild type PACAP. d Quantification of luminescence output and determination by curve-fitting of EC50 values for a biotinylated PACAP derivative (PACAP-10-BT) in the presence (green) and absence (blue) of 200 nM streptavidin (SA). e Screening for PACAP residues that tolerate modification: EC50 values for receptor activation in the absence (blue) or presence (green) of SA by PACAP-biotin conjugates as a function of sequence position (untested positions in gray). EC50 of wild-type (WT) PACAP indicated by the dashed line. f EC50 values of multiply biotinylated PACAP conjugates modified at the positions noted (N = N-terminus), in the absence (blue) and presence (green) of SA. Error bars in d–e denote SD of duplicate measurements.