Figure 4.
Reduction in X-Linked Gene Expression in Naive hPSCs Might Be Due to the Erasure of Active X Upregulation
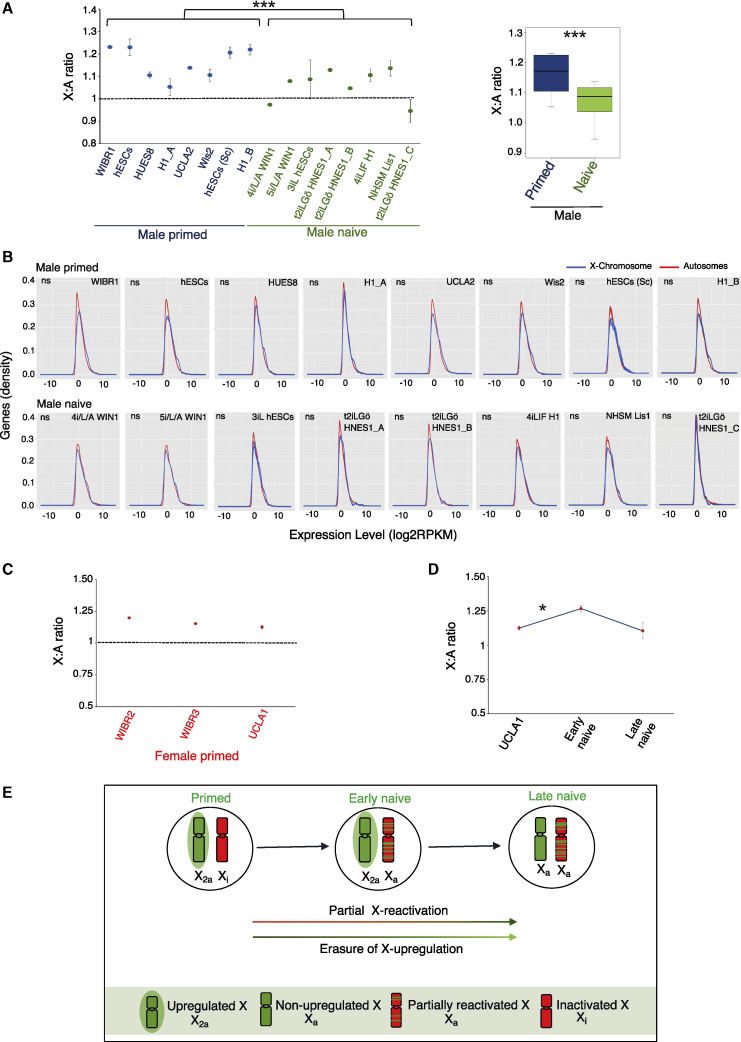
(A) Comparison of X:A ratio between male primed and naive hPSCs. p < 0.001(Student's t test). Number of independent replicates of RNA-seq dataset used were as follows. Male primed: WIBR1 (n = 1), hESCs (n = 3), HUES8 (n = 2), H1_A (n = 2), UCLA2 (n = 1), Wis2 (n = 2), hESCs (Sc) (n = 8 single cells), H1_B (n = 3). Male naive: 4i/L/A WIN1 (n = 1), 5i/L/A WIN1 (n = 1), 3iL hESCs (n = 3), t2iLGo HNES1_A (n = 3), t2iLGo HNES1_B (n = 1), 4iLIF H1 (n = 2), NHSM Lis1 (n = 2), t2iLGo HNES1_C (n = 2).
(B) Histograms representing the distribution of X-linked and autosomal gene expression for male primed and naive hPSCs with their different replicates (p > 0.05 by Kolmogorov-Smirnov test).
(C) Analysis of X:A ratio in different primed female hPSCs. Replicates of RNA-seq dataset used: WIBR2 and WIBR3 (n = 1); UCLA1 (n = 2).
(D) Comparison of X:A ratio in UCLA1 primed, early naive cells (cl4), and late naive cells (cl9 and cl12) based on two replicates of RNA-seq dataset for each. p < 0.05.
(E) Proposed model representing the X chromosome states during the conversion of primed hPSCs to the naive state.