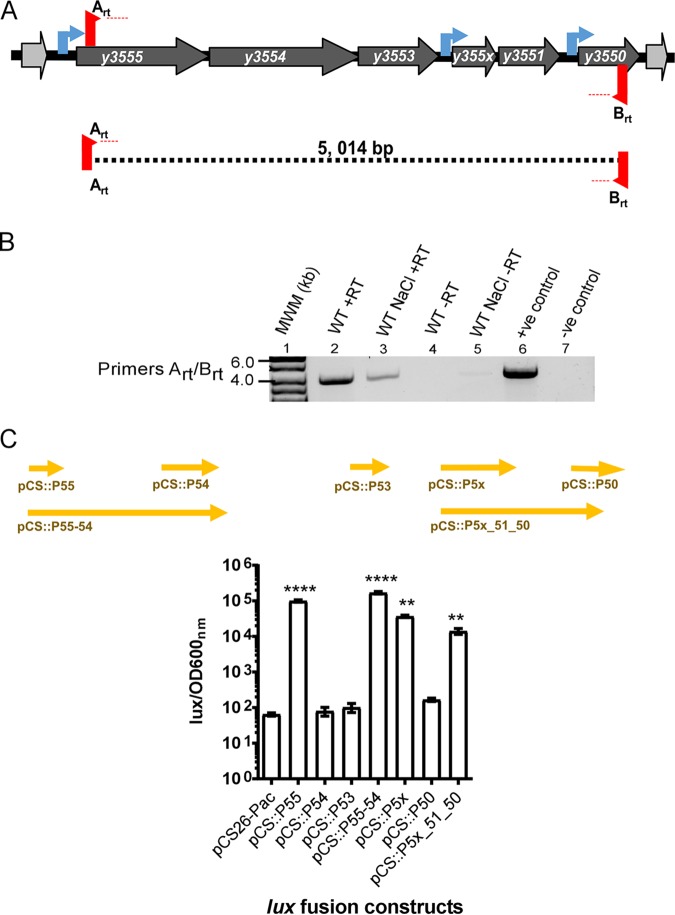
FIG 1.
The low-GC-content gene cluster is transcribed as a single polycistronic mRNA. Six genes compose the low-GC-content gene cluster. (A) The three bioinformatically predicted transcriptional units are marked by the presence of predicted promoters upstream of genes y3555, y355x, and y3550 (blue arrows). (B) Reverse transcription-PCR (RT-PCR) analysis of regions (indicated by dashed lines) spanning y3555 to y3550. RT-PCRs were performed with reverse transcriptase and DNA polymerase mix (lanes 2 and 3) or only with DNA polymerase (lanes 4 to 7). PCRs in the presence of Y. pestis KIM6+ chromosomal DNA (lane 6) or absence of template (lane 7) with the same set of primers were performed as controls. MWM, molecular weight marker; +ve, positive; -ve, negative. (C) The activity of transcriptional fusion plasmids pCS::P55, pCS::P54, pCS::P53, pCS::P55_54, pCS::P5x, pCS::P50, and pCS::P5x_51_50, carrying the transcriptional fusions of the luciferase (lux) genes and predicted promoter regions (arrows) representing predicted regulatory regions of y3555, y3554, y3553, y3555-y3554, y355x, y3550, and y355x-y3551-y3550, respectively, as well as the low-copy-number empty vector pCS26-Pac. Error bars represent mean (±standard deviation [SD]) of the results from three independent experiments. Significance determined by one-way analysis of variance (ANOVA) and a Dunnett’s posttest of lux/OD values of transcriptional fusion constructs compared to the empty vector is indicated: ****, P < 0.0001, and **, P < 0.01.